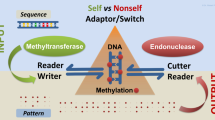
Abstract
Post-translational histone modifications and their biological effects have been described as a ‘histone code’. Independently, Barbieri used the term ‘organic code’ to describe biological codes in addition to the genetic code. He also provided the defining criteria for an organic code, but to date the histone code has not been tested against these criteria. This paper therefore investigates whether the histone code is a bona fide organic code. After introducing the use of the term ‘code’ in biology, the criteria a putative organic code such as the histone code must conform to in order to be recognised as an organic code are described. Our current knowledge of histones and their major post-translational modifications, and the specific protein binding domains that recognise and translate these into specific biological effects, is then reviewed in detail. The histone modification system is then placed in the context of an organic code and it is concluded that it fulfils all the requirements of an organic code. The marks produced on histones by processes such as acetylation and methylation act as organic signs that are translated into unique biological effects, their biological meanings. These translations are accomplished by effector proteins that consist of a binding domain that recognises a specific histone mark and a regulatory domain that mediates the biological effect. Crucially, these domains can be experimentally interchanged between different effector proteins, thus altering the rules that specify the relationships between sign and meaning. The effector proteins therefore fulfil the role of adaptor molecules.


Similar content being viewed by others
References
Agalioti, T., Chen, G., Thanos, D. (2002). Deciphering the transcriptional histone acetylation code for a human gene. Cell, 111(3), 381–392.
Allfrey, V.G., Faulkner, R., Mirsky, A.E. (1964). Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proceedings of the National Academy of Sciences of the United States of America, 51(5), 786.
Barbieri, M. (2008). Biosemiotics: a new understanding of life. Naturwissenschaften, 95(7), 577–599.
Barbieri, M. (2012). Code Biology–A New Science of Life.
Barbieri, M. (1985). The semantic theory of evolution: Chur: Harwood Academic Publishers.
Barbieri, M. (1998). The organic codes. The basic mechanism of macroevolution. Rivista di Biologia, 91(3), 481.
Barbieri, M. (2003). The organic codes: An introduction to semantic biology. Cambridge: Cambridge University Press.
Bártová, E., Krejčí, J., Harničarová, A., Galiová, G., Kozubek, S. (2008). Histone modifications and nuclear architecture: a review. Journal of Histochemistry and Cytochemistry, 56(8), 711–721.
Bell, O., Wirbelauer, C., Hild, M., Scharf, A.N.D., Schwaiger, M., MacAlpine, D.M., Zilbermann, Frédéric, van Leeuwen, F., Bell, S.P., Imhof, A., Garza, D., Peters, A.H.F.M., Schübeler, D. (2007). Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO Journal, 26(24), 4974–4984.
Berger, S.L. (2002). Histone modifications in transcriptional regulation. Current Opinion in Genetics and Development, 12(2), 142–148.
Botuyan, M.V., Lee, J., Ward, I.M., Kim, J.-E., Thompson, J.R., Chen, J., Mer, G. (2006). Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell, 127(7), 1361–1373.
Brenner, C., & Fuks, F. (2007). A methylation rendezvous: reader meets writers. Developmental Cell, 12(6), 843–844.
Brownell, J.E., Zhou, J., Ranalli, T., Kobayashi, R., Edmondson, D.G., Roth, S.Y., Allis, C.D. (1996). Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell, 84(6), 843–852.
Carrozza, M.J., Li, B., Florens, L., Suganuma, T., Swanson, S.K., Lee, K.K., Shia, W.-J., Anderson, S., Yates, J., Washburn, M.P., Workman, J.L. (2005). Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell, 123(4), 581–592.
Cavalli, G., & Paro, R. (1998). Chromo-domain proteins: linking chromatin structure to epigenetic regulation. Current Opinion in Cellular Biology, 10(3), 354–360.
Chen, C.-H., Chang, W.-F., Liu, C.-C., Su, H.-Y., Shyue, S.-K., Cheng, W.T.K., Chen, E.Y., Wu, S.-C., Du, F., Sung, L.-Y. (2012). Spatial and temporal distribution of Oct-4 and acetylated H4K5 in rabbit embryos. Reproductive BioMedicine Online, 24(4), 433–442.
Crick, F.H.C., Barnett, L., Brenner, S., Watts-Tobin, R.J. (1961). General nature of the genetic code for proteins. Nature, 192(4809), 1227–1232.
Cui, K., Zang, C., Roh, T.-Y., Schones, D.E., Childs, R.W., Peng, W., Zhao, K. (2009). Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell, 4(1), 80–93.
de la Cruz, X., Lois, S., Sánchez-Molina, S., Martínez-Balbás, M.A. (2005). Do protein motifs read the histone code? Bioessays, 27(2), 164–175.
de Napoles, M., Mermoud, J.E., Wakao, R., Tang, Y.A., Endoh, M., Appanah, R., Nesterova, T.B., Silva, J., Otte, A.P., Vidal, M., Koseki, H., Brockdorff, N. (2004). Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Developmental Cell, 7(5), 663–676.
De Santa, F., Totaro, M.G., Prosperini, E., Notarbartolo, S., Testa, G., Natoli, G. (2007). The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell, 130(6), 1083–1094.
Dey, A., Chitsaz, F., Abbasi, A., Misteli, T., Ozato, K. (2003). The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proceedings of the National Academy of Sciences of the United States of America, 100(15), 8758–8763.
Dover, J., Schneider, J., Tawiah-Boateng, M.A., Wood, A., Dean, K., Johnston, M., Shilatifard, A. (2002). Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. Journal of Biological Chemistry, 277(32), 28368–28371.
Elder, D. (1979). An epigenetic code. Differentiation, 14(1), 119–122.
Fischle, W., Wang, Y., Jacobs, S.A., Kim, Y., Allis, C.D., Khorasanizadeh, S. (2003). Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Development, 17(15), 1870–1881.
Flanagan, J.F., Mi, L.-Z., Chruszcz, M., Cymborowski, M., Clines, K.L., Kim, Y., Minor, W., Rastinejad, F., Khorasanizadeh, S. (2005). Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature, 438(7071), 1181–1185.
Gabius, H.J. (2000). Biological information transfer beyond the genetic code: the sugar code. Naturwissenschaften, 87(3), 108–121.
Gatta, R., Dolfini, D., Zambelli, F., Imbriano, C., Pavesi, G., Mantovani, R. (2011). An acetylation-monoubiquitination switch on lysine 120 of H2B. Epigenetics, 6(5), 630–637.
Gelbart, M.E., Larschan, E., Peng, S., Park, P.J., Kuroda, M.I. (2009). Drosophila MSL complex globally acetylates H4K16 on the male X chromosome for dosage compensation. Nature Structural and Molecular Biology, 16(8), 825–832.
Gifford, C.A., Ziller, M.J., Gu, H., Trapnell, C., Donaghey, J., Tsankov, A., Shalek, A.K., Kelley, D.R., Shishkin, A.A., Issner, R., Zhang, X., Coyne, M., Fostel, J.L., Holmes, L., Meldrim, J., Guttman, M., Epstein, C., Park, H., Kohlbacher, O., Rinn, J., Gnirke, A., Lander, E.S., Bernstein, B.E., Meissner, A. (2013). Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell, 153(5), 1149–1163.
Gimona, M. (2008). Protein linguistics and the modular code of the cytoskeleton. In The codes of life (pp. 189–206), Springer.
Grunstein, M. (1997). Histone acetylation in chromatin structure and transcription. Nature, 389(6649), 349–352.
Hurley, J., Lee, S., Prag, G. (2006). Ubiquitin-binding domains. Biochemical Journal, 399, 361–372.
Husnjak, K., & Dikic, I. (2012). Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual Review of Biochemistry, 81, 291–322.
Jeltsch, A., & Rathert, P. (2008). Putting the pieces together: histone H2B ubiquitylation directly stimulates histone H3K79 methylation. Chembiochem, 9(14), 2193–2195.
Jenuwein, T., & Allis, C.D. (2001). Translating the histone code. Science’s STKE, 293(5532), 1074.
Khan, A.U., & Hampsey, M. (2002). Connecting the DOTs: covalent histone modifications and the formation of silent chromatin. Trends in Genetics, 18(8), 387–389.
Kim, J., Daniel, J., Espejo, A., Lake, A., Krishna, M., Li, X., Yi, Z., Bedford, M.T. (2006). Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Reports, 7(4), 397–403.
Koche, R.P., Smith, Z.D., Adli, M., Gu, H., Ku, M., Gnirke, A., Bernstein, B.E., Meissner, A. (2011). Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell, 8(1), 96–105.
Komander, D., & Rape, M. (2012). The ubiquitin code. Annual Review of Biochemistry, 81, 203–229.
Kornberg, R.D., & Lorch, Y. (1999). Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell, 98, 285–294.
Kouzarides, T. (2007). Chromatin modifications and their function. Cell, 128(4), 693.
Kurdistani, S.K., & Grunstein, M. (2003). Histone acetylation and deacetylation in yeast. Nature Reviews Molecular Cell Biology, 4(4), 276–284.
Kurdistani, S.K., Tavazoie, S., Grunstein, M. (2004). Mapping global histone acetylation patterns to gene expression. Cell, 117(6), 721–733.
Latham, J.A., Chosed, R.J., Wang, S., Dent, S.Y.R. (2011). Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell, 146(5), 709–719.
Lee, J.-S., & Shilatifard, A. (2007). A site to remember: H3K36 methylation a mark for histone deacetylation. Mutation Research: Fundamental and Molecular Mechanisms, 618(1), 130–134.
Lee, J.-S., Shukla, A., Schneider, J., Swanson, S.K., Washburn, M.P., Florens, L., Bhaumik, S.R., Shilatifard, A. (2007). Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell, 131(6), 1084–1096.
Levenson, J.M., & Sweatt, J.D. (2005). Epigenetic mechanisms in memory formation. Nature Reviews Neuroscience, 6(2), 108–118.
Margueron, R., & Reinberg, D. (2011). The Polycomb complex PRC2 and its mark in life. Nature, 469(7330), 343–349.
Martin, C., & Yi, Z. (2005). The diverse functions of histone lysine methylation. Nature Reviews Molecular Cell Biology, 6(11), 838–849.
Maurer-Stroh, S., Dickens, N.J., Hughes-Davies, L., Kouzarides, T., Eisenhaber, F., Ponting, C.P. (2003). The Tudor domain ‘Royal Family’: tudor, plant agenet, chromo, PWWP and MBT domains. Trends in Biochemical Sciences, 28(2), 69–74.
McDaniel, I.E., Lee, J.M., Berger, M.S., Hanagami, C.K., Armstrong, J.A. (2008). Investigations of CHD1 function in transcription and development of Drosophila melanogaster. Genetics, 178(1), 583–587.
McGinty, R.K., Kim, J., Chatterjee, C., Roeder, R.G., Muir, T.W. (2008). Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature, 453(7196), 812–816.
McKittrick, E., Gafken, P.R., Ahmad, K., Henikoff, S. (2004). Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proceedings of the National Academy of Sciences of the United States of America, 101(6), 1525–1530.
Mellor, J. (2006). It takes a PHD to read the histone code. Cell, 126(1), 22–24.
Mellor, J. (2009). Linking the cell cycle to histone modifications: Dot1, G1/S, and cycling K79me2. Molecular Cell, 35(6), 729–730.
Mujtaba, S., Zeng, L., Zhou, M.M. (2007). Structure and acetyl-lysine recognition of the bromodomain. Oncogene, 26(37), 5521–5527.
Mujtaba, S., He, Y., Zeng, L., Farooq, A., Carlson, J.E., Ott, M., Verdin, E., Zhou, M.-M. (2002). Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Molecular Cell, 9(3), 575–586.
Nguyen, A.T., & Yi, Z. (2011). The diverse functions of Dot1 and H3K79 methylation. Genes & Development, 25(13), 1345–1358.
Nirenberg, M., Leder, P., Bernfield, M., Brimacombe, R., Trupin, J., Rottman, F., O’neal, C. (1965). Rna codewords and protein synthesis, vii. on the general nature of the rna code. Proceedings of the National Academy of Sciences of the United States of America, 53(5), 1161.
Org, T., Chignola, F., Hetenyi, C., Gaetani, M., Rebane, A., Liiv, I., Maran, U., Mollica, L., Bottomley, M.J., Musco, G., Peterson, P. (2008). The autoimmune regulator PHD finger binds to non-methylated histone H3K4 to activate gene expression. EMBO Reports, 9(4), 370–376.
Osley, M.A. (2004). H2B ubiquitylation: the end is in sight. BBA Gene Structural Expression, 1677(1), 74–78.
Owen, D.J., Ornaghi, P., Yang, J.-C., Lowe, N., Evans, P.R., Ballario, P., Neuhaus, D., Filetici, P., Travers, A.A. (2000). The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO Journal, 19(22), 6141–6149.
Pereira, S.L., Grayling, R.A., Lurz, R., Reeve, J.N. (1997). Archaeal nucleosomes. Proceedings of the National Academy of Sciences of the United States of America, 94(23), 12633–12637.
Peterson, C.L., & Laniel, M.-A. (2004). Histones and histone modifications. Current Biology, 14(14), 546–551.
Pokholok, D.K., Harbison, C.T., Levine, S., Cole, M., Hannett, N.M., Lee, T.I., Bell, G.W., Walker, K., Rolfe, A., Herbolsheimer, E., Zeitlinger, J., Lewitter, F., Gifford, D.K., Young, R.A. (2005). Genome-wide map of nucleosome acetylation and methylation in yeast. Cell, 122(4), 517–527.
Pray-Grant, M.G., Daniel, J.A., Schieltz, D., Yates, J.R., Grant, P.A. (2005). Chd1 chromodomain links histone H3 methylation with SAGA and SLIK-dependent acetylation. Nature, 433(7024), 434–438.
Ramakrishnan, V. (1997). Histone structure and the organization of the nucleosome. Annual Review of Biophysics and Biomolecular Structure, 26(1), 83–112.
Rice, J.C., & Allis, C.D. (2001). Histone methylation versus histone acetylation: new insights into epigenetic regulation. Current Opinion in Cellular Biology, 13(3), 263–273.
Roest, H.P., Baarends, W.M., de Wit, J., van Klaveren, J.W., Wassenaar, E., Hoogerbrugge, J.W., van Cappellen, W.A., Hoeijmakers, J.H.J., Grootegoed, J.A. (2004). The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Molecular and Cellular Biology, 24(12), 5485–5495.
Rundlett, S.E., Carmen, A.A., Suka, N., Turner, B.M., Grunstein, M. (1998). Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature, 392(6678), 831–835.
Schlesinger, D.H., Goldstein, G., Niall, H.D. (1975). Complete amino acid sequence of ubiquitin, an adenylate cyclase stimulating polypeptide probably universal in living cells. Biochemistry (Moscow), 14(10), 2214–2218.
Schneider, R., Bannister, A.J., Myers, F.A., Thorne, A.W., Crane-Robinson, C., Kouzarides, T. (2003). Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biology, 6(1), 73–77.
Schotta, G., Lachner, M., Sarma, K., Ebert, A., Sengupta, R., Reuter, G., Reinberg, D., Jenuwein, T. (2004). A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes & Development, 18(11), 1251–1262.
Schwartz, Y.B., & Pirrotta, V. (2008). Polycomb complexes and epigenetic states. Current Opinion in Cellular Biology, 20(3), 266–273.
Shahbazian, M.D., & Grunstein, M. (2007). Functions of site-specific histone acetylation and deacetylation. Annual Review of Biochemistry, 76, 75–100.
Shahbazian, M.D., Zhang, K., Grunstein, M. (2005). Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Molecular and Cellular, 19(2), 271–277.
Shi, L.H., Ai, J.S., Ouyang, Y.C., Huang, J.C., Lei, Z.L., Wang, Q., Yin, S., Han, Z.M., Sun, Q.Y., Chen, D.Y. (2008). Trichostatin A and nuclear reprogramming of cloned rabbit embryos. Journal of Animal Sciences, 86(5), 1106–1113.
Shi, X., Hong, T., Walter, K.L., Ewalt, M., Michishita, E., Hung, T., Carney, D., Pena, P., Lan, F., Kaadige, M.R., et al. (2006). ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature, 442(7098), 96–99.
Shilatifard, A. (2008). Molecular implementation and physiological roles for histone H3 lysine 4 (H2K4) methylation. Current Opinion in Cellular Biology, 20(3), 341–348.
Simic, R., Lindstrom, D.L., Tran, H.G., Roinick, K.L., Costa, P.J., Johnson, A.D., Hartzog, G.A., Arndt, K.M. (2003). Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO Journal, 22(8), 1846–1856.
Simon, J.A., & Kingston, R.E. (2009). Mechanisms of polycomb gene silencing: knowns and unknowns. Nature Reviews Molecular Cell Biology, 10(10), 697–708.
Smallwood, A., Estève, P.-O., Pradhan, S., Carey, M. (2007). Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes & Development, 21(10), 1169–1178.
Snowden, A.W., Gregory, P.D., Case, C.C., Pabo, C.O. (2002). Gene-specific targeting of H3K9 methylation is sufficient for initiating repression in vivo. Current Biology, 12(24), 2159–2166.
Söll, D., Ohtsuka, E., Jones, D.S., Lohrmann, R., Hayatsu, H., Nishimura, S., Khorana, H.G. (1965). Studies on polynucleotides, xlix. stimulation of the binding of aminoacyl-srna’s to ribosomes by ribotrinucleotides and a survey of codon assignments for 20 amino acids. Proceedings of the National Academy of Sciences of the United States of America, 54(5), 1378.
Steger, D.J., Lefterova, M.I., Ying, L., Stonestrom, A.J., Schupp, M., Zhuo, D., Vakoc, A.L., Kim, J.-E., Chen, J., Lazar, M.A., Blobel, G.A., Vakoc, C.R. (2008). DOT1L/LMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Molecular and Cellular Biology, 28(8), 2825–2839.
Strahl, B.D., & Allis, C.D. (2000). The language of covalent histone modifications. Nature, 403(6765), 41.
Strašák, L., Bártová, E., Harničarová, A., Galiová, G., Krejčí, J., Kozubek, S. (2009). H3K9 acetylation and radial chromatin positioning. Journal of Cellular Physiology, 220(1), 91–101.
Sun, Z.-W., & Allis, C.D. (2002). Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature, 418(6893), 104–108.
Swigut, T., & Wysocka, J. (2007). H3K27 demethylases, at long last. Cell, 131(1), 29–32.
Tavares, L., Dimitrova, E., Oxley, D., Webster, J., Poot, R., Demmers, J., Bezstarosti, K., Taylor, S., Ura, H., Koide, H., Wutz, A., Vidal, M., Elderkin, S., Brockdorff, N. (2012). RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell, 148(4), 664–678.
Tomkins, G.M. (1975). The metabolic code. Science, 189(4205), 760–763.
Tóth, K.F., Knoch, T.A., Wachsmuth, M., Frank-Stöhr, M., Stöhr, M., Bacher, C.P., Müller, G., Rippe, K. (2004). Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. Journal of Cell Science, 117(18), 4277–4287.
Tsai, W.-W., Wang, Z., Yiu, T.T., Akdemir, K.C., Xia, W., Winter, S., Tsai, C.-Y., Shi, X., Schwarzer, D., Plunkett, W., Aronow, B., Or, G., Fischle, W., Hung, M.-C., Patel, D.J., Barton, M.C. (2010). TRIM24 links a non-canonical histone signature to breast cancer. Nature, 468(7326), 927–932.
Turner, B.M. (2000). Histone acetylation and an epigenetic code. Bioessays, 22(9), 836–845.
Turner, B.M. (2002). Cellular memory and the histone code. Cell, 111(3), 285–291.
Vakoc, C.R., Mandat, S.A., Olenchock, B.A., Blobel, G.A. (2005). Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Molecular Cell, 19(3), 381–391.
Wang, G.G., Cai, L., Pasillas, M.P., Kamps, M.P. (2007). NUP98–NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nature Cell Biology, 9(7), 804–812.
Wang, H., Wang, L., Erdjument-Bromage, H., Vidal, M., Tempst, P., Jones, R.S., Yi, Z. (2004). Role of histone h2a ubiquitination in polycomb silencing. Nature, 431(7010), 873–878.
Wang, Z., Zang, C., Rosenfeld, J.A., Schones, D.E., Barski, A., Cuddapah, S., Cui, K., Roh, T.-Y., Peng, W., Zhang, M.Q., Zhao, K. (2008). Combinatorial patterns of histone acetylations and methylations in the human genome. Nature Genetics, 40(7), 897–903.
Wu, J., & Grunstein, M. (2000). 25 years after the nucleosome model: chromatin modifications. Trends in Biochemical Sciences, 25(12), 619–623.
Yuan, W., Mo, X., Huang, C., Liu, N., Chen, S., Zhu, B. (2011). H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. Journal of Biological Chemistry, 286(10), 7983–7989.
Zhang, Y. (2003). Transcriptional regulation by histone ubiquitination and deubiquitination. Genes & Development, 17(22), 2733–2740.
Zhu, B., Zheng, Y., Pham, A.-D., Mandal, S.S., Erdjument-Bromage, H., Tempst, P., Reinberg, D. (2005). Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Molecular Cell, 20(4), 601–611.
Acknowledgements
The authors thank Marcello Barbieri and Joachim de Beule for their boundless support and guidance. The authors acknowledge financial support from the South African National Research Foundation (NRF). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors, and therefore the NRF does not accept any liability in regard thereto.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kühn, S., Hofmeyr, JH.S. Is the “Histone Code” an Organic Code?. Biosemiotics 7, 203–222 (2014). https://doi.org/10.1007/s12304-014-9211-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12304-014-9211-2