Abstract
Although Bayesian methods are widely used in phylogenetic systematics today, the foundations of this methodology are still debated among both biologists and philosophers. The Bayesian approach to phylogenetic inference requires the assignment of prior probabilities to phylogenetic trees. As in other applications of Bayesian epistemology, the question of whether there is an objective way to assign these prior probabilities is a contested issue. This paper discusses the strategy of constraining the prior probabilities of phylogenetic trees by means of the Principal Principle. In particular, I discuss a proposal due to Velasco (Biol Philos 23:455–473, 2008) of assigning prior probabilities to tree topologies based on the Yule process. By invoking the Principal Principle I argue that prior probabilities of tree topologies should rather be assigned a weighted mixture of probability distributions based on Pinelis’ (P Roy Soc Lond B Bio 270:1425–1431, 2003) multi-rate branching process including both the Yule distribution and the uniform distribution. However, I argue that this solves the problem of the priors of phylogenetic trees only in a weak form.



Similar content being viewed by others
Notes
While the question of whether the MCMC analysis is sufficiently thorough is of high importance to practitioners, this issue is set aside in this more conceptually minded paper.
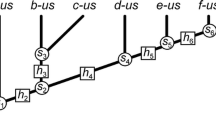
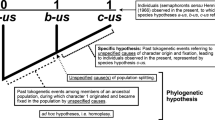
If not explicitly stated otherwise, I refer to a tree topology when using the term ‘tree’ or ‘phylogenetic tree’ in this paper.
Hence, the Yule process is also known as ‘pure birth process’ or just a ‘birth process’.
Pinelis refers to the uniform distribution on tree topologies as the ‘proportional-to-distinguishable-arrangements’ (PDA) model.
Since the set of states S is finite these two sums are finite.
For a very sceptical view on Eldredge and Gould’s idea of punctuated equilibrium, see Dennett (1995).
An alternative stochastic process inducing the uniform distribution on tree topologies is the ‘explosive radiation process’ suggested by Steel and McKenzie (2001). In the explosive radiation process ‘stabilization’ (rather than quasi-stabilization) is certain to occur for every species that reach a certain age. That is, in the explosive radiation process a species older than a certain age does not give birth to any new species nor does it change in any way.
Idealizations that involve deliberate distortions are sometimes referred to as ‘Galilean idealizations’ (McMullin 1985).
For more details on the hierarchical Bayesian approach in phylogenetics, see Yang (2006), p. 123.
In fact, applying the full Bayesian approach to some stochastic processes can be very restrictive. Take the Yule process as one, admittedly extreme, example. Applying the full Bayesian approach to this process yields the Yule distribution for any possible prior probability distribution of the model parameter since for any (constant) choice of this parameter the Yule process induces the Yule distribution.
References
Aldous DJ (1996) Probability distributions on cladograms. In: Aldous DJ, Permantle R (eds) Random discrete structures. Springer, New York, pp 1–18
Aldous DJ (2001) Stochastic models and descriptive statistics for phylogenetic trees. Statist Sci 16:23–34
Alfaro ME, Holder MT (2006) The posterior and the prior in bayesian phylogenetics. Annu Rev Ecol Evol Syst 37:19–42
Dennett DC (1995) Darwin’s dangerous idea: evolution and the meanings of life. Simon and Schuster, New York
Earman J (1992) Bayes or bust? A critical examination of bayesian confirmation theory. MIT Press, Cambridge
Edwards WH, Lindman H, Savage LJ (1963) Bayesian statistical inference for psychological research. Psychol Rev 70:193–242
Eldredge N, Gould SJ (1972) Punctuated equilibria: an alternative to phyletic gradualism. In: Schopf TJM (ed) Models in paleobiology. Freeman Cooper, San Francisco, pp 82–115
Felsenstein J (2004) Inferring phylogenies. Sinauer, Sunderland
Gillies D (2000) Philosophical theories of probability. Routledge, London
Guyer C, Slowinski J (1993) Adaptive radiation and the topology of large phylogenies. Evolution 47:253–263
Heard SB (1992) Patterns in tree balance among cladistic, phenetic, and randomly generated phylogenetic trees. Evolution 46:1818–1826
Howson C, Urbach P (2006) Scientific reasoning: the bayesian approach, 3rd edn. Open Court, Chicago
Huelsenbeck JP, Rannala B (2004) Frequentist properties of bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol 53:904–913
Huelsenbeck JP, Ronquist F (2001) MR BAYES: bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Jaynes ET (1983) In: Rosenkrantz R (ed) Papers on probability, statistics, and statistical physics. Reidel, Dordrecht
Keynes JM (1921) A treatise on probability. MacMillan, New York
Levins R (1966) The strategy of model building in population biology. Am Sci 54:421–431
Lewis D (1980) A subjectivist’s guide to objective chance. In: Jeffrey R (ed) Studies in inductive logic and probability vol II. University of California Press, Berkeley, pp 263–293
Maddison W, Slatkin M (1991) Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution 45:1184–1197
McMullin E (1985) Galilean idealization. Stud Hist Philos Sci 16:247–273
Mellor DH (2005) Probability: a philosophical introduction. Routledge, London
Pinelis I (2003) Evolutionary models of phylogenetic trees. P Roy Soc Lond B Bio 270:1425–1431
Rannala B, Yang Z (1996) Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J Mol Evol 43:304–311
Simmons MP, Pickett KM, Miya M (2004) How meaningful are bayesian support values? Mol Biol Evol 21:188–199
Steel M, McKenzie A (2001) Properties of phylogenetic trees generated by Yuletype speciation models. Math Biosci 170:91–112
Sterelny K (2007) Dawkins vs. Gould: survival of the fittest. Icon Books, Thriplow
Suppes P (1966) A bayesian approach to the paradoxes of confirmation. In: Hintikka J, Suppes P (eds) Aspects of inductive logic. North-Holland, Amsterdam, pp 198–207
Velasco JD (2008) The prior probabilities of phylogenetic trees. Biol Philos 23:455–473
Weisberg M (2006) Robustness analysis. Philos Sci 73:730–742
Williamson J (2007) Motivating objective bayesianism: from empirical constraints to objective probabilities. In: Harper WL, Wheeler GR (eds) Probability and inference: essays in honor of Henry E. Kyburg Jr. College Publications, London, pp 155–183
Wimsatt WC (1981) Robustness, reliability, and overdetermination. In: Brewer M, Collins B (eds) Scientific enquiry and the social sciences. Jossey-Boss, San Francisco, pp 124–163
Yang Z (2006) Computational molecular evolution. Oxford University Press, Oxford
Yule GU (1924) A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS. P Roy Soc Lond B Bio 213:21–87
Acknowledgments
I would like to thank Jason Alexander, Roman Frigg and Elliott Sober as well as two anonymous reviewers for very helpful comments on earlier drafts of this paper.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Autzen, B. Constraining prior probabilities of phylogenetic trees. Biol Philos 26, 567–581 (2011). https://doi.org/10.1007/s10539-011-9253-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10539-011-9253-7