Towards mining scientific discourse using argumentation schemes
Abstract
The dominant approach to argument mining has been to treat it as a machine learning problem based upon superficial text features, and to treat the relationships between arguments as either support or attack. However, accurately summarizing argumentation in scientific research articles requires a deeper understanding of the text and a richer model of relationships between arguments. First, this paper presents an argumentation scheme-based approach to mining a class of biomedical research articles. Argumentation schemes implemented as logic programs are formulated in terms of semantic predicates that could be obtained from a text by use of biomedical/biological natural language processing tools. The logic programs can be used to extract the underlying scheme name, premises, and implicit or explicit conclusion of an argument. Then this paper explores how arguments in a research article occur within a narrative of scientific discovery, how they are related to each other, and some implications.
1.Introduction
The dominant approach to argument (or argumentation) mining [6,14,29] has been to treat it as a machine learning problem requiring use only of superficial text features, enabling researchers to adopt methods that have been applied successfully to other natural language processing tasks. That approach has been successful for a variety of applications such as identifying reasons given for opinions in social media, or automatic assessment of student essay quality. However accurately summarizing argumentation in scientific research articles may require a deeper understanding of the text.
There are a number of problems with mining arguments in scientific documents using only superficial text features rather than at a semantic level [9,11]. Argument components may not occur in contiguity in the text. In fact, the content of an argument may be widely separated or the content of two arguments may be interleaved at the text level. Furthermore scientific text often contains enthymemes, i.e. arguments with some implicit premises or an implicit conclusion. Interpretation of enthymemes may require use of the preceding discourse context (including inferred conclusions of other arguments), presumed shared knowledge of the author and audience, as well as constraints of the underlying argumentation scheme [10].
Although human-level understanding of natural language text is currently beyond the state of the art, we propose a semantics-informed approach to extracting individual arguments from biomedical research articles on human genetic variants with adverse health effects. This paper describes how argumentation schemes implemented as logic programs in Prolog [2] could be used to extract individual arguments in that genre. The schemes are formulated in terms of semantic predicates that could be obtained from a text by use of BioNLP (biomedical/biological natural language processing) tools. However, extracting individual arguments is not sufficient for understanding the argumentation in a scientific research article. Instead, it is necessary to model the argumentation structure, or inter-argument relationships, in the article. As a step towards that, this paper explores how arguments in one research article are related to its narrative discourse structure, and how they are related to each other. Examining this narrative of scientific discovery suggests possible contextual constraints on semantics-informed argument mining and illustrates that a richer model of inter-argument relationships is needed.
2.Argumentation schemes
Argumentation schemes are abstract descriptions of acceptable, possibly defeasible, arguments used in conversation as well as in formal genres such as legal and scientific text [37]. This section describes our proposal to mine arguments using argumentation schemes implemented as logic programs. We present seven argumentation schemes that we defined in Prolog after analyzing the arguments in the first eight paragraphs of a ten-paragraph “Results/Discussion” section of a biomedical research article on the putative cause of a genetic disorder [13]. The article [35] was selected from the open-access CRAFT11 corpus, which has been annotated by other researchers for purposes of biomedical text mining, but not for argument mining [1,36]. The schemes were implemented in terms of a small set of domain-specific semantic predicates (Fig. 1) that could in theory be automatically extracted from a text by BioNLP tools. We expect these schemes to be applicable to the large and growing body of research articles on genetic variants with adverse health effects.
Fig. 1.
Domain Predicate Definitions.
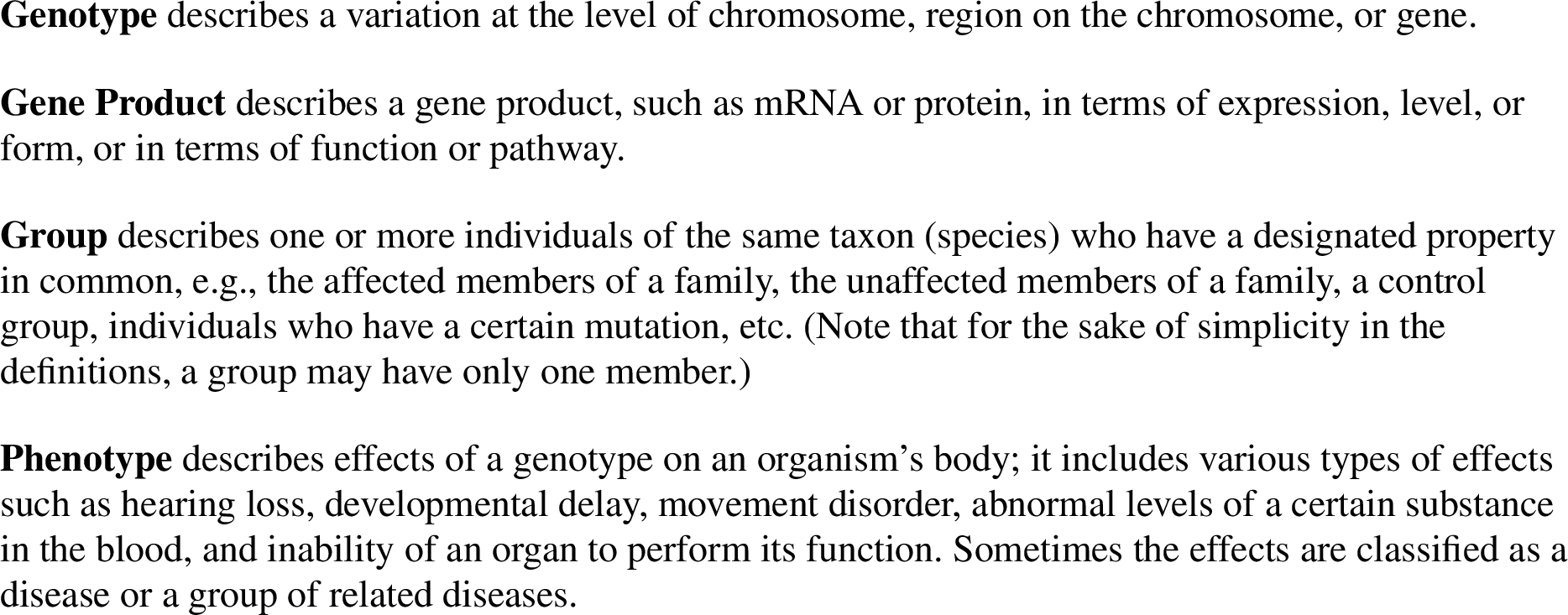
Fig. 2.
Agreement.
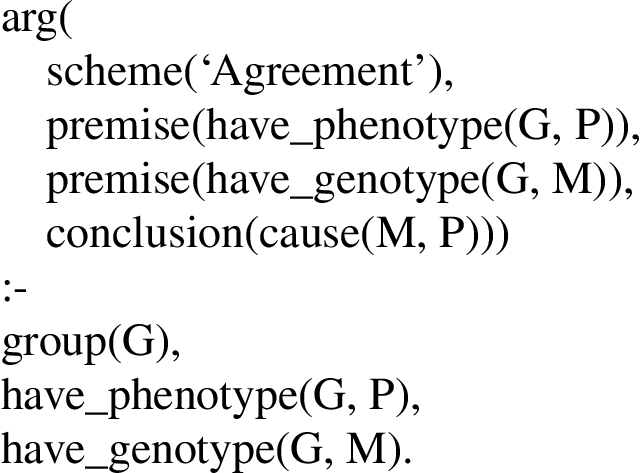
Fig. 3.
Difference.

The schemes in Fig. 2 and Fig. 3 are specializations of Mill’s Method of Agreement and Method of Difference [16], respectively, which were formulated to describe scientific reasoning in the nineteenth century. (For readers who are not familiar with Prolog notation, see Section 3 for more information about the representation of the schemes.) Our Agreement scheme can be paraphrased as follows: If a group has an abnormal genotype G and abnormal phenotype P then G may be the cause of P (in that group). Our Difference scheme can be paraphrased as follows: If a group has an abnormal genotype G and abnormal phenotype P and it is known that a second group has neither G nor P, then G may be the cause of P (in the first group). The knot operator used in the implementation of Difference and some other schemes can be read as “know not”, to differentiate it from Prolog’s negation-as-failure operator. Also, note that the conclusions of the schemes given in this article are not asserted with complete certainty. The corresponding arguments in the source text range in force from ‘plausible hypothesis’ to ‘fairly certain conclusion’.
Fig. 4.
Failed Agreement (No Effect).

Fig. 5.
Eliminate Difference.

The scheme in Fig. 4, dubbed Failed Agreement (No Effect), is also related to Mill’s Method of Agreement. It can be paraphrased as If a group has an abnormal genotype G and is known not to have abnormal phenotype P then it is not likely that G is a cause of P. The Eliminate Difference scheme shown in Fig. 5 is related to Difference (Fig. 3). In Difference, the presence/absence of a potential causal agent is correlated with the presence/absence of an observed potential effect. In Eliminate Difference, the presence/absence of the potential causal agent A is implied, based upon the knowledge that A is the difference in the presence of AB and of B. This scheme can be paraphrased as If a group has an abnormal genotype AB and abnormal phenotype P and a second group has genotype B (without A) and is known not to have P then A may be the cause of P (in the first group). The schemes shown in Figs 2 through 5 also could be considered more specific versions of a Correlation to Cause argument scheme [37]. However, there is no requirement for correlation in the statistical sense. Also, these and the following schemes presuppose two principles of modern biology: first, that genotype may influence phenotype but not conversely, and second, that biological mechanisms operate in a consistent manner in different individuals.
The Analogy scheme shown in Fig. 6 bears some resemblance to the preceding schemes, as well as to Argument by Analogy, e.g. as described in [37]. Our version of Analogy can be paraphrased as If a group has abnormal genotype G and abnormal phenotype P and G may be the cause of P, and a second group has abnormal genotype G’ similar to G and abnormal phenotype P’ similar to P, then G’ may be the cause of P’. Our version differs from the more general definition of Argument by Analogy in licensing a conclusion about a causal relationship.
Fig. 6.
Analogy.

Fig. 7.
Consistent Explanation.
The Consistent Explanation scheme shown in Fig. 7 can be paraphrased as If a group has an abnormal genotype G, abnormal gene product Prot, and abnormal phenotype P, and G produces Prot, and Prot is associated with P, and G may cause P, then if a second group has an abnormal genotype G’ similar to G and abnormal phenotype P’ similar to P, and G’ produces Prot and Prot is associated with P’, then G’ may be the cause of P’. In other words, if a certain causal mechanism explains how G causes P (via Prot), and an analogous causal mechanism could explain how G’ could cause P’ (via the same Prot), then G’ may be the cause of P’. Thus, this rule involves analogies as well as the above two principles of modern biology.
The Difference Consistent Explanation scheme shown in Fig. 8 combines aspects of Difference (Fig. 3) and the previous scheme, namely, if a certain causal mechanism explains how, in one group, abnormal genotype G causes abnormal phenotype P (via abnormal product Prot), and there is a second group that is known not to have G, Prot, nor P, then G may be the cause of P.
Fig. 8.
Difference Consistent Explanation.

The above schemes have been evaluated informally as follows. Previously, we drafted a catalog which included some similar argumentation scheme definitions based upon arguments that we had found in several other articles on genetic variants with adverse health effects [11]. Unlike the definitions given here, the schemes were defined in terms of non-domain-specific concepts. A small study showed that, given these definitions, other researchers could successfully identify the argument schemes of arguments in excerpts of articles of this sort [11]. After implementing the seven schemes described here based upon the author’s solo analysis of the CRAFT article [13], the article was re-analyzed by a team consisting of the author and two graduate student assistants (one a doctoral student with a background in genetics, the other a computer science major with an undergraduate degree in philosophy), and the seven types of arguments were re-analyzed. In the process of reanalyzing the article, the author rewrote the catalog of scheme definitions in terms of the domain-specific concepts (genotype, phenotype, etc.) implemented in the logic programs. Then another small study was done to see if, given the revised catalog and excerpts of the CRAFT article containing arguments with implicit conclusions, other researchers could reliably identify the premises and argumentation scheme for five types of argument [12]. Although mainly successful, the study revealed some confusion between schemes such as Analogy and Consistent Explanation, pointing to the need to better explain those schemes in a catalog for use by human annotators. An updated catalog of 15 argumentation schemes is now available.22
The Prolog implementation of the rules was tested using a manually created knowledge base [13]. The next section describes how such a knowledge base could be created automatically and how the rules could be used in an argument mining process.
3.Argument mining process
This section summarizes our proposed approach to mining individual arguments using argumentation schemes like those described in Figs 2–8 in the preceding section. The first step would be to apply BioNLP tools to a source text to create a knowledge base (KB). Named entity recognition tools such as ABNER [32] or MutationFinder [5] could be used to recognize expressions referring to semantic class names such as genes, mutations, proteins, and phenotypes. Domain-specific relations in the argumentation schemes such as have_phenotype and have_genotype could be extracted from the text using relation extraction tools such as OpenMutationMinder [26] and DiMeX [20]. Also, a certain amount of domain knowledge would be required, e.g., for the relations similar and difference, which could be acquired from a domain ontology or domain experts. After a KB has been created, the argument scheme rules described in the preceding section would be applied to the KB to recognize the premises, conclusion, and argumentation scheme of each argument in the text.
Fig. 9.
Argument Mining Example.
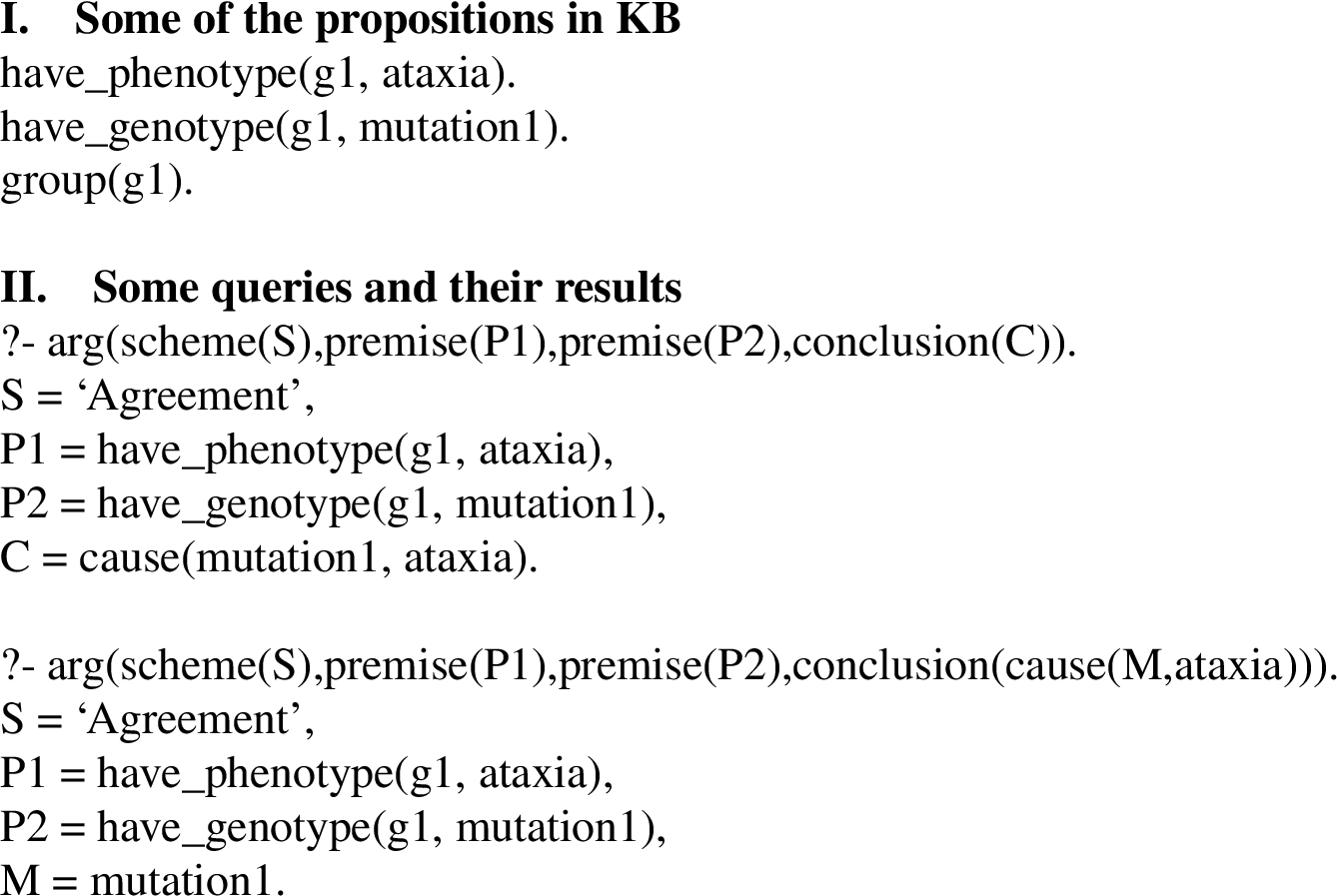
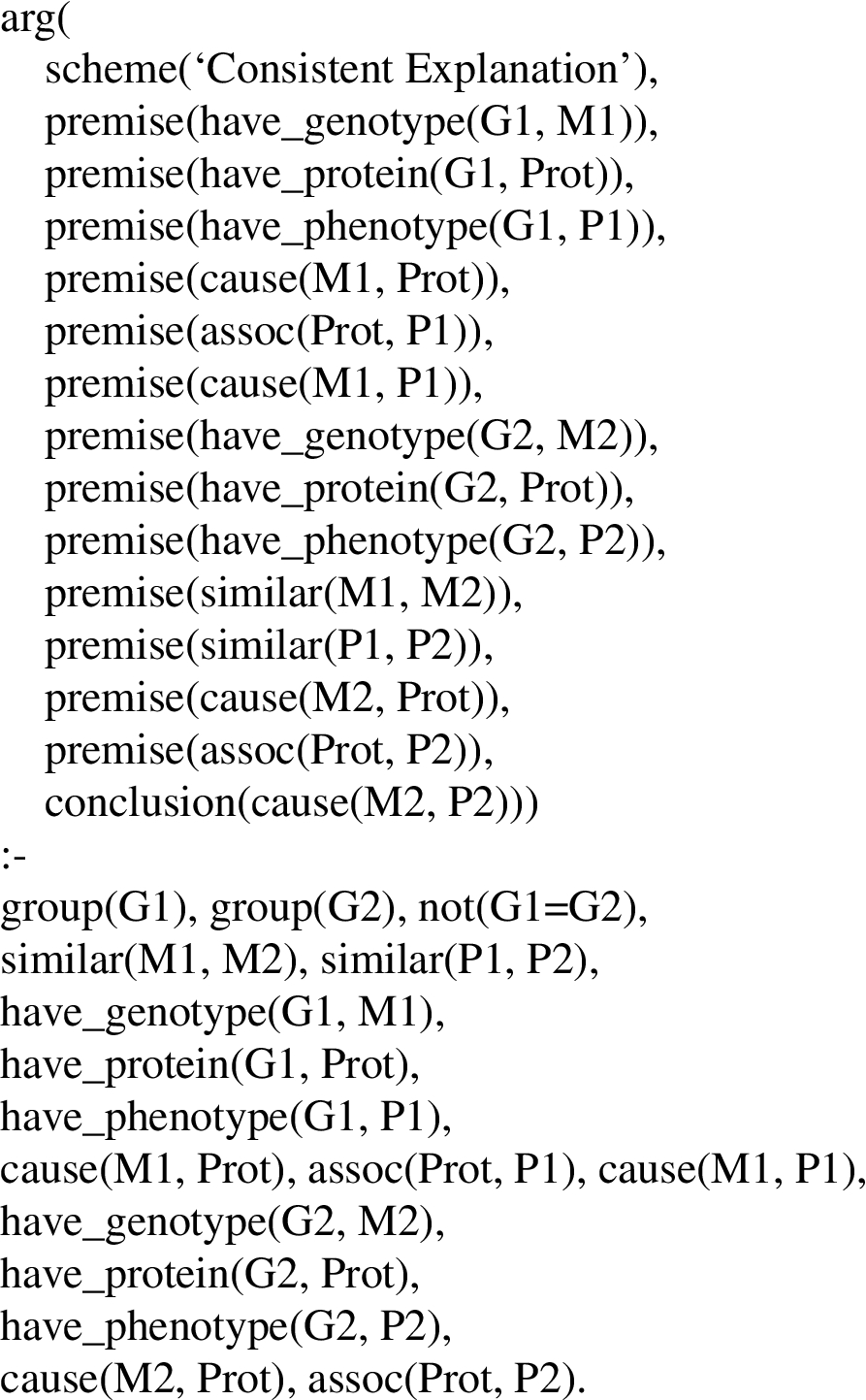
To illustrate this process, suppose that after creation of a KB by application of BioNLP tools to a biology article, the KB contains many Prolog propositions, including the three shown in Fig. 9, part I. One way to make use of the argument schemes shown in Figs 2–8 is to pose the Prolog query,
Prolog could derive an argument as an answer to the query as follows. For those unfamiliar with Prolog notation, note that the argumentation schemes shown in Figs 2–8 consist of two parts, separated by ‘:-’. The top part (to the left of ‘:-’) is a goal to be proven and the bottom part (to the right of ‘:-’) contains a list of propositions all of which must be proven in order to prove the goal. Both the goal and the list of propositions may contain variables, indicated by terms beginning with an upper case letter. In the process of proving a goal the Prolog theorem prover may replace variables with constants. In order to answer the above query, the theorem prover will apply schemes whose goal matches the query such as the Agreement argumentation scheme (Fig. 2). In the bottom part of the Agreement rule, group(G), have_phenotype(G,P), and have_genotype(G,M) would match group(
Since a KB typically would contain many more propositions extracted from an article than the three shown in the example, the above query might derive additional arguments using other argumentation schemes and/or other facts. Also, depending upon how the system is intended to be used, more specific queries could be posed, e.g., find arguments for causes of ataxia:
Prolog could then derive the argument described above, where M is replaced with mutation1.
A possible limitation of this approach is the necessity to first extract semantic relations. However there is considerable effort in the BioNLP community to develop relation extraction tools for other purposes. Another possible limitation is the cost of manually deriving rules for topics not covered by the current rules. As noted above, the rules are specializations of argumentation schemes that have been described in research on argumentation in general and on biomedical argumentation. Thus it is plausible that the effort to create rules for other topics by specialization will not be significantly higher than the cost of formulating the current rule set. Also, since the size of the targeted literature is large and constantly growing, the benefits may outweigh the cost. (However, see the Discussion section on how scheme rules might be acquired in the future.)
4.Discourse context of arguments
Having proposed an approach to mining individual arguments, the next step of our research is to investigate how the arguments are related to other aspects of discourse structure, and how the arguments are related to each other. This section describes how they are related to discourse structure, and inter-argument relationships are discussed in the next section. Previous computation-oriented investigations of discourse in the natural sciences have addressed automatic classification of text segments, e.g., discourse coherence relations in corpora such as BioDRB [28] and BioCause [23], argumentative zones [34], and activities in a scientific investigation (CoreSC) [19]. None of those annotation schemes treat arguments in the sense described in the preceding sections.
Table 1
Discourse Context of Arguments
| Discourse Segment | D. Subsegment | Argument Component (Arg. ID, Scheme) |
| Report Observation | Premise (Arg. 1), Premise(Arg. 3) | |
| Report Experiment 1 | Result | Premise (Arg. 1) |
| Conclusion (implicit) | Conclusion (Arg. 1, Agreement) | |
| Premise (Arg. 2) | ||
| Report Experiment 2 | Previous Research | Premise (Arg. 2) |
| Hypothesis (implicit) | Conclusion (Arg. 2, Analogy) | |
| Result | Premise (Arg. 3) | |
| Conclusion (implicit) | Conclusion (Arg. 3, Agreement) | |
| Report Experiment 3 | Previous Research | Premise (Arg. 4) |
| Result | Premise (Arg. 4) | |
| Conclusion (implicit) | Conclusion (Arg. 4, Consistent Explanation) | |
| Premise (Arg. 4s) | ||
| Summary | Conclusion (implicit) | Conclusion (Arg. 4s, Agreement) |
| Premise (Arg. 5) | ||
| Report Experiment 4 | Background Knowledge | Premise (Arg. 5) |
| Hypothesis | Conclusion (Arg. 5, Analogy) | |
| Result | Premise (Arg. 6) | |
| Premise (Arg. 7) | ||
| Conclusion (implicit) | Conclusion (Arg. 6, Agreement) | |
| Report Experiment 5 | Results | Premise (Arg. 7) |
| Conclusion (implicit) | Conclusion (Arg. 7, Difference) | |
| Report Experiment 6 | Result | Premise (Arg. 8) |
| Conclusion (implicit) | Conclusion (Arg. 8, Difference) | |
| Report Experiment 7 | Result | Premise (Arg. 9) |
| Conclusion (implicit) | Conclusion (Arg. 9, Difference) | |
| Summary | Result | Premise (Arg. 10) |
| Conclusion | Conclusion (Arg. 10, Difference) | |
| Premise (Arg. 12) | ||
| Discussion | Inference | Conclusion (Arg. 11, Failed Agreement) |
| Premise (Arg. 12) | ||
| Background Knowledge | Premise (Arg. 11) | |
| Inference (implicit) | Conclusion (Arg. 12, Eliminate Difference) | |
| Conclusion | Conclusion (Arg. 13, Consistent Explanation) | |
| Result (Experiment 3) | Premise (Arg. 13) | |
| Background Knowledge | Premise (Arg. 13) | |
| Conclusion | Conclusion (Arg. 14, Difference Consistent Explanation) | |
| Result | Premise (Arg. 14) |
The first eight paragraphs of the Results/Discussion section of the CRAFT article reports on a logical and temporal sequence of experiments. Arguments are given in the context of this narrative, i.e. a report of the scientific investigation. Table 1 shows our ad hoc analysis of the narrative using descriptive terms similar to those of the argumentative zone and CoreSC systems. The section begins with a description of the fortuitous discovery of an inherited ataxia-like disorder in some mice bred in the authors’ lab (labeled Report Observation). Then the authors describe a sequence of experiments (each labeled Report Experiment). The experiments are described in Table 1 in terms of Previous Research, Background Knowledge, Hypothesis, Result, and/or Conclusion. (Parts of a reported experiment that did not contain argument components, such as Method, are not indicated.)
The first three experiments were designed to reveal the genetic variant responsible for the ataxia-like mouse disorder. At the end of the report of the first three experiments, the article summarizes findings about known causes of ataxia in mice (labeled Summary). The goal of the subsequent experiments was to discover any related genetic variants causing a similar disorder in humans. The section ends with the authors’ final conclusion about the cause of ataxia in the individuals discussed in Experiments 4 through 7 (labeled Summary) and further arguments in support of it as a plausible cause of ataxia in humans in general (labeled Discussion).
Table 1 also shows the location of premises and conclusions of each argument and the name of its argumentation scheme in the context of this narrative structure. It can be seen that argument boundaries do not coincide with narrative segments. Also note that in most cases an argument’s conclusion is implicit. Moreover, the implicit conclusions of Arguments 1 and 4 function as premises of subsequent arguments. (The frequent occurrence of implicit conclusions poses quite a challenge for approaches based only on superficial text features.) To support our interpretation of some of these implicit conclusions, note the conclusions of Arguments 3, 4 and 5 are stated in the Abstract section, the conclusion of Arguments 3 and 4 is also stated in the Author Summary section, and less specific versions of conclusions of Arguments 4s and 12, 13, and 14 are stated in the article’s title. When conclusions are explicitly given, they are preceded by premises in Argument 5 and 10, and followed by premises in Arguments 11, 13, and 14. Thus, it is unclear from study of this article alone whether or not the ordering of premises and conclusion can be exploited for argument mining.
This analysis raises interesting questions for future research. First, are uses of certain argumentation schemes more likely to occur in certain discourse contexts? In the article, Analogy is used twice (and only) to argue for a hypothesis. (Perhaps there are normative constraints in this genre on the appropriate use of schemes for certain discourse goals.) Second, are certain sequences of uses of argumentation schemes more likely to occur? In the Discussion segment, for example, the sequence Failed Agreement (Argument 11) and Eliminate Difference (Argument 12) functions as an argumentative unit. The above observations could be verified by statistical analyses of corpora, and if true, could augment the semantic method we have proposed for extracting individual arguments. Another possibility is that the location in the narrative could be used as a constraint on argument scheme recognition: Although argument content is not in one-to-one correspondence with discourse boundaries, in cases where multiple argument scheme rules match, a heuristic strategy of preferring local content might be applied.
5.A narrative of discovery
Table 2 shows that to describe this article a richer model of relationships between arguments is needed than simple support-attack relationships represented in current argument mining approaches, e.g., [4,27,33]. To explain the analysis in Table 2, first, note that the conclusion of all of the arguments is of the form cause (CausalAgent, Effect, Population). (Population specifies the population to which the causal statement applies, e.g., the affected mice bred in the authors’ lab, human family members affected with a certain disorder, or humans in general. Also, note that Population is not always explicitly specified in the text and was not included in the conclusions of the implemented argumentation schemes.) The conclusion of Argument 2 refines the CausalAgent in the conclusion of Argument 1. Similarly, Arguments 3 and 12 refine the CausalAgent in the conclusion of Arguments 2 and 10, respectively. Employing the Consistent Explanation scheme, Argument 4 addresses a critical question that could be raised to challenge the preceding argument, namely, whether the genetic variant is biologically plausible as a cause of the mouse disorder. The conclusion of Argument 4s aggregates the Population of Argument 3 with other mice discussed in previous work, and broadens the CausalAgent of Argument 3 to include the related mutations associated with the other mice.
Table 2
Dialectical structure
| Arg. | Conclusion/Relationship to other argument | Related Arg. |
| 1 | cause( 6qE1_lesion, ataxia_like, lab_mice). | |
| 2 | cause( hom_del_Itpr1, ataxia_like, lab_mice). | 1 |
| Refines causal agent from lesion on chromosome 6qE1 to homozygous deletion in Itpr1 gene on 6qE1. | ||
| 3 | cause(Itpr1Δ18/Δ18, ataxia_like, lab_mice). | 2 |
| Refines causal agent from homozygous deletion in Itpr1 to a specific deletion on Itpr1 (Itpr1Δ18/Δ18). | ||
| 4 | Same as 3. | 3 |
| Responds to CQ. | ||
| 4s | cause( Itpr1_deletions, ataxia_like, {targeted, opt, lab_mice}). | 3 |
| Aggregates population of lab_mice with two other groups of mice. | ||
| Broadens Itpr1Δ18/Δ18 to Itpr1_deletions. | ||
| 5 | cause( del_ITPR1, SCA15, humans). | 4s |
| Analogous conclusion. (SCA15 is a type of ataxia.) | ||
| 6 | cause( del_ITPR1_SUMF1, SCA15, AUS1). | 5 |
| Broadens causal agent from ITPR1 deletion to deletion in ITPR1_SUMF1 region. | ||
| Restricts population from humans to affected AUS1 family members. | ||
| 7 | Same as 6. | 6 |
| Agrees. | ||
| 8 | Same as 6. | 6 |
| Agrees | ||
| 9 | cause( del_ITPR1_SUMF1, SCA15_like, {H33, H27}). | 6 |
| Broadens effect from SCA15 to SCA15_like. | ||
| Changes population from affected AUS1 to affected H33 and H27 family members. | ||
| 10 | cause( del_ITPR1_SUMF1, SCA15, {AUS1,H33, H27}). | 9 and 6 |
| Aggregates population of affected members of H33 and H27 with affected members of AUS1. | ||
| Refines effect from SCA15_like to SCA15. | ||
| 11 | not cause( del_SUMF1, SCA15, humans). | 10 |
| Rejects part of causal agent (not SUMF1 gene). | ||
| Broadens population to humans. | ||
| 12 | cause(del_ITPR1, SCA15, humans). | 10 |
| Refines causal agent from ITPR1_SUMF1 deletion to ITPR1 deletion | ||
| 13 | Same as 12. | 12 |
| Responds to CQ. | ||
| 14 | Same as 12. | 12 |
| Responds to CQ |
At this point, the narrative shifts focus to the cause of a similar disorder in humans. The relationship of Argument 5 to Argument 4s is that their conclusions are analogous in terms of CausalAgent (mouse Itpr1 gene vs. human ITPR1 gene), Effect (mouse ataxia-like disorders vs. human ataxia-like disorders), and Population (mouse vs. human). Argument 6 both broadens the CausalAgent in the conclusion of Argument 5, while restricting the Population in the conclusion of Argument 5. The conclusions of Arguments 7 and 8 simply agree with the conclusion of Argument 6. On the other hand, the conclusion of Argument 9 broadens the Effect and Population in the conclusion of Argument 6. Argument 10 aggregates the Populations in the conclusions of Argument 9 and 6 and refines the Effect of Argument 9. Argument 11 rejects part of the CausalAgent in the conclusion of Argument 10, while broadening its Population to humans in general. Argument 12 refines the CausalAgent in the conclusion of Argument 10. Arguments 13 and 14 provide responses to a critical question of Argument 12, namely, is the causal relation biologically plausible?
It would be misleading to reduce these relationships to simply support or attack in an automated summarization of the content. As described in research on formal dialogue games for use by software agents, the discovery dialogue [22] has a key feature in common with the scientific article that we analyzed. In a discovery dialogue, the goal is not to try to prove or disprove a given claim, but to discover something not previously known. In this genre when reporting on the discovery of the genetic cause of phenotypes in a population, the moves of the discovery dialogue can be summarized not only as asserting, supporting, or attacking a conclusion or responding to a critical question, but also as
Refining/Broadening/Rejecting CausalAgent or Effect,
Asserting an analogous conclusion, or
Broadening/Aggregating/Restricting Population.
It should be noted that it may also be necessary to model the relative strengths of arguments according to field-specific criteria, e.g., that evidence from a mouse model is not as strong as evidence from humans.
6.Related work
Argumentative zoning (AZ) annotation schemes were developed for automatically classifying the sentences of scientific articles (in computational linguistics and chemistry) in terms of their contribution of new knowledge to a field [34]. For example, categories include CONTRAST (negatively contrasting competitors’ knowledge claims to the author’s), BACKGROUND (generally accepted background knowledge), and OWN (the author’s new knowledge claim). An extension of AZ for genetics research articles includes categories such as MTH (methodology), RSL (experimental results), and IMP (implications of experimental results or previous work) [24]. The CoreSC (Core Scientific Concepts) annotation scheme was developed for automatic classification of sentences in terms of the components of a scientific investigation: Hypothesis, Motivation, Goal, Object, Background, Method, Experiment, Model, Observation, Result and Conclusion [19].
Two BioNLP corpora, BioDRB [28] and BioCause [23], have been annotated with discourse coherence relations similar to those defined in Rhetorical Structure Theory (RST) [21]. However, annotation of discourse coherence relations is not sufficient for argument mining since rhetorical structure may diverge from argument structure, and coherence relation definitions do not characterize distinctions among argumentation schemes [9,11,27].
In seminal work on argument mining, Mochales and Moens [25] developed a multi-stage approach applied to the Araucaria corpus [30] and a legal corpus. Machine learning was used to (1) classify sentences as part of an argument or not, (2) determine the boundaries of each argument, and (3) classify the sentences in an argument as premise or conclusion. Then in the final stage, manually constructed context-free grammar rules were used to detect relationships between arguments. On the other hand, Cabrio and Villata [4] used an approach based on calculating textual entailment [7] to detect support and attack relations between arguments in a corpus of on-line dialogues stating user opinions.
Feng and Hirst [8] investigated the problem of argumentation scheme recognition in the Araucaria corpus, given text labeled as the premises and conclusions of arguments. They built classifiers to recognize occurrences of the five most frequently used schemes in the corpus: Argument from example, Argument from cause to effect, Practical reasoning, Argument from consequences, and Argument from verbal classification. In addition to general features applicable to all five schemes, they used scheme-specific phrases such as ‘for example’ and punctuation cues.
In contrast to the above machine learning approaches, Saint-Dizier [31] used manually-derived rules encoded in a logic programming language for automatic identification of arguments giving reasons for a conclusion in instructional texts or opinion texts. However, unlike in our approach, the rules are based on syntactic patterns and lexical features.
Lawrence and Reed [18] investigated the problem of recognizing the “proposition type” of argument components in the AIFdb corpus [17]. In this corpus, the premises of an argument are subclassified according to type, e.g., the two premises of Practical Reasoning are labeled as Goal or GoalPlan. Thus, the proposition type provides a limited amount of semantic information. Using the labeling of premise type, conclusion, and argument scheme of text in the corpus, Lawrence and Reed built classifiers to recognize individual premises and conclusions based upon text features. They then experimented with identifying instances of schemes in a corpus of arguments extracted from a 19th century philosophy text. After classifying the proposition type of premises and conclusion of each text segment in the corpus, groupings of segments that could belong to the same argumentation scheme were identified. Missing components were assumed to indicate enthymemes.
7.Discussion
We have proposed an alternative to text-feature-based approaches to argument mining. A semantics-informed approach can avoid various problems in mining scientific articles faced by those approaches, e.g., that argument components may be conveyed through non-contiguous or overlapping text segments of varying granularity, a sparcity of discourse cues marking argument components, and the occurrence of enthymemes. Aside from the problems noted above, current approaches require a large corpus of annotated arguments, and there is currently no such corpus for biological/biomedical research articles. No doubt that is due to the difficulty (and expense) of analysis of argumentation in a highly specialized scientific domain.
Rather than annotate a scientific corpus at the text level, in [12] we propose creation of a semantically annotated corpus of arguments in a partially automated, two-step process. The first step would be to identify the entities and relations in the text. This could be done manually or using BioNLP tools. The second step would be to manually document the arguments in terms of the entities and relations annotated in the first step, i.e., at a propositional level. (However, a boot-strapping approach might be possible, where arguments might be provisionally annotated by use of argument mining rules, and then edited by human annotators.) Such a corpus could be used to validate argument mining results. Another possible use of the corpus, once created, would be to investigate automatic acquisition of semantics-informed argument mining rules through use of inductive logic programming (ILP) [3,15]. ILP might be used to incorporate contextual constraints into the argumentation schemes as well.
In addition to illustrating the implementation of argumentation schemes in terms of propositions that could be extracted by semantic tools, we investigated the relationship of arguments to narrative discourse structure. Although as we showed there is not a simple one-to-one relationship between argument boundaries and narrative segments, we suggest that contextual information provided by current tools that analyze scientific discourse structure could augment the use of argumentation schemes. Finally, we showed that the dialectical structure of argumentation in this type of article is more complex than has been assumed and suggest that it resembles in some respects the discovery dialogue type. In order to automatically generate useful summaries of argumentation, it may be advisable not only to extract individual arguments but to situate them in the narrative and dialectical structure.
Notes
2 Available at https://github.com/greennl/BIO-Arg.
Acknowledgements
The checking of argument analyses was done with the help of graduate students, Michael Branon and Bishwa Giri, who were supported by a University of North Carolina Greenboro 2016 Summer Faculty Excellence Research Grant.
References
[1] | M. Bada, M. Eckert, D. Evans et al., Concept annotation in the CRAFT corpus, BMC Bioinformatics 13: ((2012) ), 161. |
[2] | I. Bratko, Prolog Programming for Artificial Intelligence, 3rd edn, Addison-Wesley, Harlow, England, (2001) . |
[3] | I. Bratko and S. Muggleton, Applications of inductive logic programming, Communications of the ACM 38: (11) ((1995) ), 65–70. doi:10.1145/219717.219771. |
[4] | E. Cabrio and S. Villata, Generating abstract arguments: A natural language approach, in: Computational Models of Argument: Proceedings of COMMA 2012, B. Verheij, S. Szeider and S. Woltran, eds, IOS Press, Amsterdam, (2012) , pp. 454–461. |
[5] | J.G. Caporaso, W.A. Baumgartner Jr., D.A. Randolph, K.B. Cohen and H.L. Mutation, Finder: A high-performance system for extracting point mutation mentions from text, Bioinformatics 23: ((2007) ), 1862–1865. doi:10.1093/bioinformatics/btm235. |
[6] | C. Cardie et al. (eds), Second Workshop on Argumentation Mining. North American Conference of the Association for Computational Linguistics, Denver (CO), (2015) . |
[7] | I. Dagan, B. Dolan, B. Magnini and D. Roth, Recognizing textual entailment: Rationale, evaluation, and approaches, Natural Language Engineering 15: (4) ((2009) ), i–xvii. |
[8] | V.W. Feng and G. Hirst, Classifying arguments by scheme, in: Proceedings of the 49th Annual Meeting of the Association for Computational Linguistics, Portland (OR), (2011) , pp. 987–996. |
[9] | N. Green, Annotating evidence-based argumentation in biomedical text, in: Proceedings of the 2015 International Workshop on Biomedical and Health Informatics, IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2015), Washington (DC), Nov. 9–12, 2015, IEEE Computer Society Press, (2015) . |
[10] | N.L. Green, Representation of argumentation in text with rhetorical structure theory. Argumentation 24: (2) ((2010) ), 181–196. doi:10.1007/s10503-009-9169-4. |
[11] | N.L. Green, Identifying argumentation schemes in genetics research articles, in: Proceedings of the Second Workshop on Argumentation Mining, North American Conference of the Association for Computational Linguistics, Denver (CO), (2015) . |
[12] | N.L. Green, Manual identification of arguments with implicit conclusions using semantic rules for argument mining, in: Proceedings of the 4th Workshop on Argument Mining, Empirical Models of Natural Language Processing (EMNLP 2017), Copenhagan, (2017) . |
[13] | N.L. Green, Implementing Argumentation Schemes as Logic Programs. In: Workshop on Computational Models of Natural Argument (CMNA 2016). International Joint Conference on Artificial Intelligence, New York. CEUR Workshop Vol-1876. |
[14] | N.L. Green et al. (eds), First workshop on argumentation mining, in: Association for Computational Linguistics, Baltimore (MD), (2014) . doi:10.3115/v1/W14-21. |
[15] | S. Gulwani, J. Hernandez-Orallo, E. Kitzelmann, S. Muggleton, U. Schmid and B. Zorn, Inductive programming meets the real world, Communications of the ACM 8: (11) ((2015) ), 90–99. |
[16] | M. Jenicek and D. Hitchcock, Logic and Critical Thinking in Medicine, American Medical Association Press, (2005) . |
[17] | J. Lawrence, F. Bex, C. Reed and M. Snaith, AIFdb: Infrastructure for the argument web, in: Proc. of the Fourth International Conference on Computational Models of Argument (COMMA 2012), (2012) , pp. 515–516. |
[18] | J. Lawrence and C. Reed, Argument mining using argumentation scheme structures, in: Computational Models of Argument: Proceedings of COMMA 2016, P. Baroni et al., eds, IOS Press, Amsterdam, pp. 379–390. |
[19] | M. Liakata et al., Automatic recognition of conceptualization zones in scientific articles and two life science applications, Bioinformatics 28: (7) ((2012) ). doi:10.1093/bioinformatics/bts071. |
[20] | A.S. Mahmood, T.J. Wu, R. Mazumder and K. Vijay-Shanker, DiMeX: A text mining system for mutation-disease association extraction, PLoS One ((2016) . |
[21] | W.C. Mann and S.A. Thompson, Rhetorical structure theory: Towards a functional theory of text organization, Text 8: (3), 243–281. |
[22] | P. McBurney and S. Parsons, Chance discovery using dialectical argumentation, in: New Frontiers in Artificial Intelligence, T. Terano et al., eds, Lecture Notes in Artificial Intelligence, v.2253, Springer Verlag, Berlin, (2001) , pp. 414–424. doi:10.1007/3-540-45548-5_57. |
[23] | C. Mihaila, T. Ohta, S. Pyysalo and S. Ananiadou, BioCause: Annotating and analysing causality in the biomedical domain, BMC Bioinformatics 14: ((2013) ), 2. |
[24] | Y. Mizuta, A. Korhonen, T. Mullen and N. Collier, Zone analysis in biology articles as a basis for information extraction, International Journal of Medical Informatics 75: (6) ((2005) ), 468–487. doi:10.1016/j.ijmedinf.2005.06.013. |
[25] | R. Mochales and M. Moens, Argumentation mining, Artificial Intelligence and Law 19: ((2011) ), 1–22. doi:10.1007/s10506-010-9104-x. |
[26] | N. Naderi and W. Witte, Automated extraction and semantic analysis of mutation impacts from the biomedical literature, BMC Genomics 13: (Suppl 4) ((2012) ), 510. |
[27] | A. Peldszus and M. Stede, Rhetorical structure and argumentation structure in monologue text, in: Proc. of the 3rd Workshop on Argumentation Mining, ACL, Berlin, (2016) . |
[28] | R. Prasad, S. McRoy, N. Frid, A. Joshi and H. Yu, The Biomedical Discourse Relation Bank, BMC Bioinformatics 12: ((2011) ), 188. |
[29] | C. Reed et al. (eds), Third Workshop on Argumentation Mining, Association for Computational Linguistics, Berlin, (2016) . |
[30] | C. Reed, R. Mochales-Palau, M. Moens and D. Milward, Language resources for studying argument, in: Proceedings of the 6th Conference on Language Resources and Evaluation (LREC2008), ELRA, pp. 91–100. |
[31] | P. Saint-Dizier, Process natural language arguments with the <TextCoop> platform, Argument and Computation 3: (1) ((2012) ), 49–82. doi:10.1080/19462166.2012.663539. |
[32] | B. Settles, ABNER: An open source tool for automatically tagging genes, proteins, and other entity names in text, Bioinformatics 21: (14) ((2005) ), 3191–3192. doi:10.1093/bioinformatics/bti475. |
[33] | C. Stab and R. Gurevych, Annotating argument components and relations in persuasive essays, in: Proceedings of COLING 2014, pp. 1501–1510. |
[34] | S. Teufel, The Structure of Scientific Articles: Applications to Citation Indexing and Summarization, CSLI Publications, Stanford, CA, (2010) . |
[35] | J. van de Leemput, J. Chandran, M. Knight et al., Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans, PLoS Genetics e108: ((2007) ), 1076–1082. |
[36] | K. Verspoor, K.B. Cohen, A. Lanfranchi et al., A corpus of full-text journal articles is a robust evaluation tool for revealing differences in performance of biomedical natural language processing tools, BMC Bioinformatics 13: ((2012) ), 207. doi:10.1186/1471-2105-13-207. |
[37] | D. Walton, C. Reed and F. Macagno, Argumentation Schemes, Cambridge University Press, (2008) . |



