Altered choroid plexus gene expression in major depressive disorder
- 1Molecular and Behavioral Neuroscience Institute, University of Michigan, Ann Arbor, MI, USA
- 2Department of Psychiatry, University of Michigan, Ann Arbor, MI, USA
- 3Psychiatry and Human Behavior, University of California - Irvine, Irvine, CA, USA
- 4Department of Psychiatry and Behavioral Sciences, Stanford University, Palo Alto, CA, USA
- 5Department of Psychiatry, Weill Cornell Medical College, Cornell University, Ithaca, NY, USA
- 6HudsonAlpha Institute for Biotechnology, Huntsville, AL, USA
Given the emergent interest in biomarkers for mood disorders, we assessed gene expression in the choroid plexus (CP), the region that produces cerebrospinal fluid (CSF), in individuals with major depressive disorder (MDD). Genes that are expressed in the CP can be secreted into the CSF and may be potential biomarker candidates. Given that we have previously shown that fibroblast growth factor family members are differentially expressed in post-mortem brain of subjects with MDD and the CP is a known source of growth factors in the brain, we posed the question whether growth factor dysregulation would be found in the CP of subjects with MDD. We performed laser capture microscopy of the CP at the level of the hippocampus in subjects with MDD and psychiatrically normal controls. We then extracted, amplified, labeled, and hybridized the cRNA to Illumina BeadChips to assess gene expression. In controls, the most highly abundant known transcript was transthyretin. Moreover, half of the 14 most highly expressed transcripts in controls encode ribosomal proteins. Using BeadStudio software, we identified 169 transcripts differentially expressed (p < 0.05) between control and MDD samples. Using pathway analysis we noted that the top network altered in subjects with MDD included multiple members of the transforming growth factor-beta (TGFβ) pathway. Quantitative real-time PCR (qRT-PCR) confirmed downregulation of several transcripts that interact with the extracellular matrix in subjects with MDD. These results suggest that there may be an altered cytoskeleton in the CP in MDD subjects that may lead to a disrupted blood-CSF-brain barrier.
Introduction
The choroid plexus (CP) is composed primarily of capillary beds, the pia mater and a large number of epithelial cells. The CP produces cerebrospinal fluid (CSF), removes byproducts, plays a role in neuroendocrine signaling and provides structural support for the brain (Skipor and Thiery, 2008; Wolburg and Paulus, 2010). Yet to date, no human post-mortem gene expression studies of the CP have been published.
As the CP is found largely in the lateral ventricles, it is often co-dissected with surrounding brain tissue. This is particularly evident in dissections of the human hippocampus, a brain structure known to respond to antidepressants and play a role in neurogenesis (Cameron and McKay, 2001; Mallei et al., 2002; Duman, 2004; Bachis et al., 2008). Given the proximity of the CP in the lateral ventricle to the hippocampus and the ability of the CP to secrete proteins into the CSF that can act on the hippocampus, it is surprising that this structure has not been previously studied in individuals with mood disorders. The CP has, however, been shown to exhibit alterations following chronic stress in rats, a model known to induce depression-like behavior (Sathyanesan et al., 2012).
It is also important to consider the disease relevance of the source material. We do not know, for example, whether proteins found in the CSF are related to gene expression in the CP or are derived from other brain areas. Previous studies have reported differences in growth factor levels in the CSF (Kahl et al., 2009; Kiec-Wilk et al., 2010; Takebayashi et al., 2010). However, conflicting evidence exists that differential expression of a growth factor in gray matter did not translate to detectable differences in expression or levels in the blood or CSF (Lanz et al., 2012). This suggests that growth factors in the CSF might arise from many different tissue sources, including the CP.
Although nothing is known about gene expression in the CP in MDD subjects, a significant amount of literature has previously focused on the role of the hippocampus. For example, there is evidence for altered gene expression in the human post-mortem hippocampus in individuals with MDD (Sequeira et al., 2007, 2009). However, gene expression studies should be interpreted with caution, as hippocampal dissections also likely contain CP. In the present study, we used laser capture microscopy to selectively isolate the CP from control and MDD post-mortem human brain prior to assessment of gene expression using microarrays. An unbiased approach was also used to determine the top network and functions that may be altered in the CP in subjects with MDD. A subset of mRNAs differentially expressed by microarray analysis was then validated by qRT-PCR. This study was the first to determine gene expression in the CP in normal controls. We also observed differences in gene expression between normal controls and individuals with MDD.
Methods and Materials
Post-Mortem Tissue Quality and Demographics
All human brains were obtained from the Brain Donor Program at the University of California—Irvine, and the studies were approved by both the University of California—Irvine and the University of Michigan Institutional Review Boards. Written informed consent was obtained from the next-of-kin of the deceased. Information regarding diagnosis, treatment and other clinically relevant variables were obtained from medical records, the coroner's investigation and family interviews. Table 1 shows a list of subject demographics for controls and individuals with MDD used in this study. Six subjects per group were used for microarray analyses, and an additional four subjects per group were added for the qRT-PCR validation study. No significant differences in age, gender, post-mortem interval or brain pH were detected between groups. All subjects also had an agonal factor score (Gustafsson et al., 2003) of 0. Previous studies found that AFS and brain pH are the strongest factors influencing gene expression (Li et al., 2004). Therefore, all brains used in this study had a pH greater than 6.6. The brains were removed at the time of autopsy, cooled to 4°C and then sliced into 0.75 cm coronal slabs which were then immediately stored at −80°C to preserve integrity (Jones et al., 1992). The slabs were then manually dissected while kept frozen at −80°C by trained staff into various brain region blocks, including the hippocampus which was used for this study.
mRNA in Situ Hybridization
Previously dissected blocks from the post-mortem human hippocampus were sectioned (−20°C) at 10 μm and stored at −80°C. One section was placed onto a SuperFrostPlus slide (Thermo FisherScientific, Waltham, MA) and stored at −80°C until processing. Every 50th section was selected for mRNA in situ hybridization throughout the hippocampus, as previously described (Lopez-Figueroa et al., 2004). Briefly, slides were fixed in 4% paraformaldehyde, rinsed in 2X SSC, incubated in an acetic anhydride/triethanolamine solution and dehydrated. Sections were then hybridized with 35S-UTP and 35S-CTP cRNA probes overnight at 55°C. The next day, slides were washed, rinsed in increasing stringency of SSC, dehydrated and exposed to Kodak Biomax MR film (Eastman Kodak, Rochester, NY). Exposure time was empirically determined using test slides to visualize the region of interest. The cRNA probes were synthesized from human cDNA cloned in-house. The GAD67 (NM_000817, 738-964) cRNA probe was used to determine the anatomical level of the human hippocampus, and the transthyretin (TTR; NM_000371, 477-701) cRNA probe was used to visualize the CP. In situ hybridization autoradiograms were then digitized and scanned using the ScanMaker 1000XL Pro Flatbed Scanner (Microtek, Carson, CA) and SilverFast Ai Imaging software (LaserSoft Imaging, Sarasota, FL).
Laser Capture Microscopy (LCM)
The hippocampus was identified using GAD67 mRNA in situ hybridization, see Figure 1. The hippocampus was identified based on the C-like shape of the dentate gyrus. Transthyretin has been shown from mouse studies to be highly abundant in the CP and can be used as a reliable mRNA marker of this tissue (Marques et al., 2011). Sections containing the CP were processed through the following dehydration protocol: room temperature for 30 s, 75% ethanol for 30 s, dH20 for 30 s, 75% alcohol for 30 s, two 95% alcohol washes for 30 s, 100% alcohol for 30 s, two xylene washes for 5 min each, and left to air dry for 20 min. Laser capture microscopy of the CP was performed at the level of the caudal hippocampus (levels 4–6, based on Amaral and Insuasti, 1990). Two field-of-views of the CP from one section was captured using the AutoPix LCM system (Arcturus, Mountain View, CA) onto CapSure Macro LCM caps (Arcturus, Mountain View, CA). Laser settings were set to 70 mW and 1.5 ms. Figure 2 shows a low magnification image (4X) of the CP before and after capture (area circled in red). The large blood vessels were not captured resulting in a sample of primarily epithelial cells.
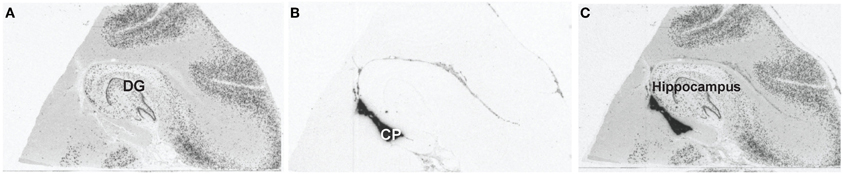
Figure 1. Identifying the choroid plexus by mRNA in situ hybridization. (A) Representative image of GAD67 gene expression to identify the hippocampal level for each subject. The dentate gyrus is the C-shaped structure in this image depicting level 5 of the hippocampus. (B) Representative image of TTR gene expression used to identify the choroid plexus for each subject. The choroid plexus is the dark region ventral to the hippocampus. (C) Representative image of GAD67 and TTR overlays.
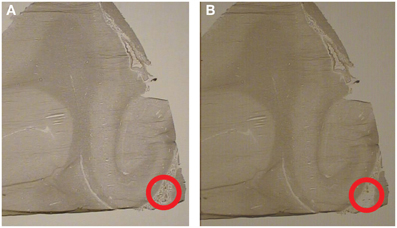
Figure 2. Laser-capture microscopy images from the same subject before and after capture. Representative images from the laser capture microscope (4X). The hippocampus is to the left and the choroid plexus within the lateral ventricle is located inside the circle. (A) Representative image of the choroid plexus before laser capture. (B) Representative image of the choroid plexus after laser capture. Notice that the choroid plexus has been removed from the area inside the circle.
RNA Isolation and Amplification
Total RNA was extracted from adherent cells (on LCM caps) using the PicoPure RNA isolation kit (Arcturus, Mountain View, CA). Cell extracts were stored at −80°C between RNA extraction and final isolation. An optional DNAse treatment was used in the isolation process. RNA was eluted in a final volume of 11 μl. Quality and quantity of cRNA was determined on the Agilent 2100 Bioanalyzer using Picochip kits (Agilent, Palo Alto, CA). The quality of 18S and 28S peaks were determined as previously described (Bernard et al., 2011). For an average area of 2.7 mm2, we obtained an average concentration of 1 ng/μl and RNA quality was determined as previously described (Kerman et al., 2012). This process yielded an average of 11 ng of total RNA. The total isolated RNA was then amplified by the RiboAmp Plus 1.5-round RNA Amplification kit (Molecular Devices, Sunnyvale, CA). The final biotin-labeled cRNA was generated with the Bioarray High Yield RNA transcription kit (ENZO life sciences, Plymouth Meeting, PA). The quantity was determined on the Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA). After amplification, the total biotin-labeled cRNA was an average of 5 μg per sample.
Microarray Analysis
Equal amounts (750 ng) of amplified total RNA sample from each human sample was hybridized to HumanHT-12v4.0 Expression BeadChips and scanned on the BeadStation system (Illumina Inc., San Diego, CA) following the manufacturer's instructions (Turner et al., 2011). Each Beadchip provides coverage of more than 47,000 probes. The microarray data was quantile normalized in BeadStudio using Illumina's error model (Illumina Inc., San Diego, CA). Fold-changes and p-values were calculated from the average array signal. Data were also analyzed by Ingenuity Pathways Analysis Software v8.0 (Ingenuity©Systems, Redwood City, CA). The Functional Analysis tool identified biological functions and/or diseases that were most significant to the data set. Molecules from the dataset that had a fold-change cutoff of 1.1, a p-value less than 0.05 and were associated with biological functions and/or diseases in Ingenuity's® Knowledge Base were considered for the analysis. To follow-up on the top pathway identified by Ingenuity Pathway Analysis, we evaluated gene ontology using Cytoscape v2.8.2 with a BinGO plug-in.
qRT-PCR
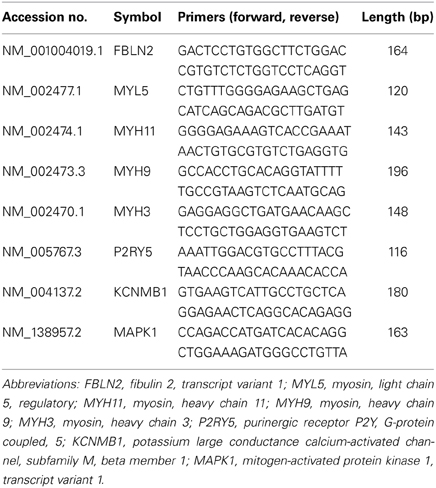
Table 2 shows a list of primers used for qRT-PCR validation. Amplified cRNA (1 μ g) was reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) in a total reaction volume of 20 μ l. cDNA (1 μl) was used as the template for real-time PCR assays with a MyiQ real-time PCR system (Bio-Rad Laboratories). The quantitative PCR was conducted in duplicate using iQ SYBR Green Supermix, according to the manufacturer's instructions (Bio-Rad Laboratories). Relative expression of the gene of interest was normalized to β-actin expression in each sample. It is important to note that β-actin did not differ between controls and individuals with MDD. The expression level of the gene of interest was evaluated using the 2−(ΔΔCt) method and values for each gene were expressed as fold-changes (Livak and Schmittgen, 2001). The PCR product quality was monitored using post-PCR melt-curve analysis at the end of the amplification cycles.
Statistical Analyses
For qRT-PCR, a Student's t-test was performed using SPSS (IBM, Armonk, NY). For pathway analysis, a right-tailed Fisher's exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that data set is due to chance alone. For gene ontology, Benjamini & Hochberg false discovery rate (FDR) correction was applied to the dataset.
Results
Controls
The most highly expressed known transcript in the human post-mortem CP was transthyretin (TTR). The primary role of TTR is to transport thyroxine (T4) and retinol in the brain (Fleming et al., 2009). However, TTR also has proteolytic activity (e.g., neuropeptide Y and β-amyloid). TTR protein levels had previously been associated with depression, such that low levels of TTR have been correlated with high suicidal ideation and low 5-HIAA (Sullivan et al., 2006). A previous study also found TTR to be downregulated in the CSF of individuals with MDD (Ditzen et al., 2011). However, TTR gene expression was not altered in the CP in MDD subjects in our study.
Supplementary Table 1 lists the 11,506 transcripts that were significantly detected across all samples in the control CP. Notably, half of the 14 mostly highly expressed transcripts were transcripts that encode ribosomal proteins. However, none of these transcripts were altered in subjects with MDD.
Subjects with MDD
In general, there were 169 transcripts differentially expressed between MDD subjects and controls, see Supplementary Table 2 for the complete list. The majority of these transcripts (75%) were downregulated with only a small percentage of transcripts upregulated (25%). The fact that the majority of the transcripts were downregulated is similar to what we have observed in the hippocampus in subjects with MDD (unpublished observations).
When Ingenuity pathway analysis was performed on the dataset, the top five significant functions were connective tissue disorder, dermatological diseases, developmental disorder, genetic disorder, and metabolic disease. The top two significant pathways were hepatic fibrosis and calcium signaling. The top network focused on transforming growth factor-beta (TGFβ). As shown in Figure 3, all of the transcripts associated with the TGFβ network were downregulated in MDD subjects compared to controls. Gene ontology analysis confirmed that growth factor binding (corrected p-value: 0.00885) was the most significant function in this dataset. Indeed, seven members of the TGFβ network validated by this analysis. TGFβ signaling is known to play a role in cytoskeletal dynamics and actin reorganization (Baghdassarian et al., 1993; Gagelin et al., 1995). Since TGFβ is associated with various components of the extracellular matrix, we decided to interrogate transcripts with high fold-changes related to structural support and integrity, some of which are part of the TGFβ pathway.
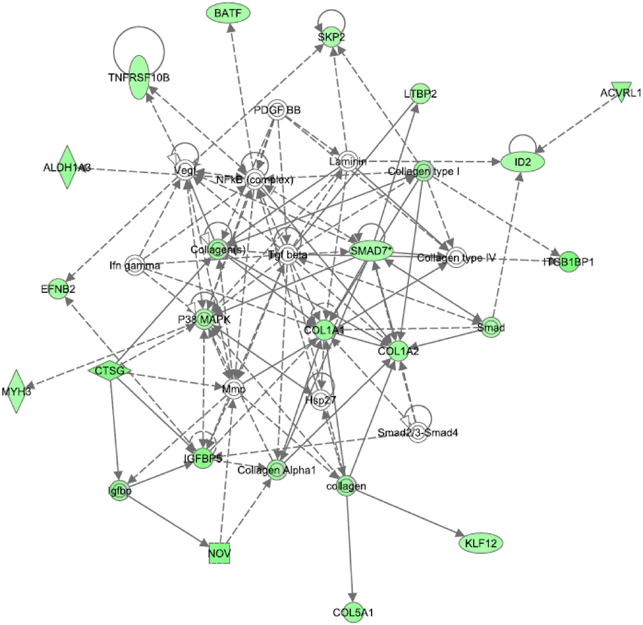
Figure 3. Top network identified from Ingenuity pathway analysis in the choroid plexus of MDD subjects. TGFβ is at the center of the top network dysregulated in MDDs. All of the transcripts in this network were downregulated in MDDs.
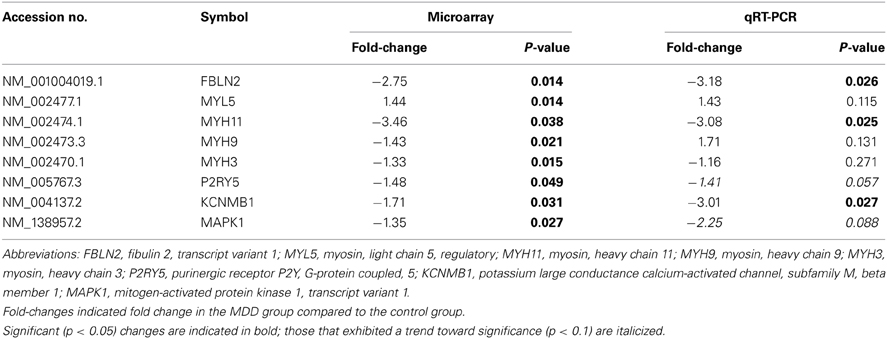
We selected eight transcripts for real-time PCR validation, see Table 3. Since we can predict the direction of change based on the microarray results, we performed one-tail t-tests for qRT-PCR. Three transcripts were confirmed by qRT-PCR and two transcripts exhibited non-significant trends in the same direction as the microarray results. We will describe these five transcripts in the next few paragraphs. Fibulin 2 (FBLN2) was significantly decreased in subjects with MDD compared to controls. FBLN2 is a secreted glycoprotein that is produced by epithelial cells and can bind calcium. In terms of function, FBLN2 interacts with many other proteins, including the laminins and integrins, to stabilize the extracellular matrix (Zhang et al., 1996; de Vega et al., 2009). It is possible that downregulation of this transcript may lead to destabilization of the extracellular matrix.
A myosin heavy chain transcript and a calcium-activated potassium channel were also altered in individuals with MDD. Myosin heavy chain 11 (MYH11) was significantly decreased in MDD subjects compared to controls. This myosin motor can bind actin and play a role in ATP hydrolysis (Renard et al., 2011; Armstrong et al., 2012). Thus, the actin cytoskeleton might be altered in subjects with MDD. The calcium-activated and voltage-dependent potassium channel β1 subunit (KCNMβ1) was also significantly decreased in individuals with MDD. Although not much is known about the role of this large conductance channel in the brain, it has been linked to cardiovascular disease and hypertension in the periphery (Grimm et al., 2009). However, it is plausible that this channel also may play a role in filtration, as total gene expression in the kidney is similar to that of the CP suggesting that the two tissues may share similar functions (Sathyanesan et al., 2012). This particular subunit of the channel is also the regulatory subunit of the channel and may alter surface expression of the receptor (Toro et al., 2006).
There were also non-significant trends for mitogen-activated protein kinase 1 (MAPK1) and the purinergic receptor, P2YR5, to be altered in MDD subjects. MAPK is part of the TGFβ network (see Figure 2) and is known to play a role in tyrosine kinase receptor signaling. MAPK is also known to bind to and phosphorylate cytoskeletal proteins (Veeranna et al., 2000). Moreover, P2YR5 (aka LPAR6) plays a role in endothelial cell morphology and may be involved in actin reorganization, as it is a receptor for lysophosphatidic acid (Ishii et al., 2009). It is also possible that this receptor may regulate vascular permeability in the CP. In summary, all five of the transcripts from the qRT-PCR experiments were downregulated in subjects with MDD. Taken together, these results suggest that there may be an altered extracellular matrix or cytoskeleton in MDD subjects.
Discussion
This study is the first to assess gene expression in the CP in MDD subjects, as well as in psychiatrically normal controls. Moreover, two salient pieces of information have emerged from this study. First, transcripts that encode ribosomal proteins were highly expressed in the CP in controls. Secondly, multiple transcripts that interact with the extracellular matrix and cytoskeleton were downregulated in individuals with MDD. Thus, there may be a disrupted CP in subjects with MDD.
In comparing our control data in humans to a previously published mouse study that used light microscopy dissection of the CP and microarrays, there was reasonable agreement among highly expressed transcripts (Marques et al., 2011). For example, there was a 25% overlap between our top 12 transcripts and their top 12 transcripts (excluding predicted sequences), including several members involved in energy demand (e.g., cox4i1 and atp5b). Moreover, this finding is in agreement with the high number of mitochondria known to exist in the CP (Cornford et al., 1997; Johanson et al., 2011). Interestingly, the P-glycoprotein pump (ABCB1) was not significantly detected in control tissue. This is likely because ABCB1 is located in the vascular endothelium near the epithelial cells of the CP (Mercier et al., 2004; Gazzin et al., 2008; Roberts et al., 2008; Kratzer et al., 2013).
This study identified transcripts that were differentially expressed between controls and MDD subjects in the CP. We did not find alterations in growth factors in the CP of MDD brains. However, several transcripts involved in calcium signaling were altered in the CP. Calcium signaling may represent another interesting area of therapeutic investigation in depression. We identified the TGFβ network to be identified by Ingenuity Pathway Analysis, and the significance of this pathway was confirmed by gene ontology analysis. The finding that the TGFβ network was altered suggests that less anti-inflammatory molecules may also be present in the CP of individuals with MDD. Further studies should be designed to follow-up on TGFβ signaling in subjects with MDD. Interestingly, MAPK1, a member of the TGFβ pathway, was the only one of the five interesting transcripts by qRT-PCR previously known to be altered in MDD brains. Single nucleotide polymorphisms in MAPK1 have been associated with treatment resistance and remission in depression (Calati et al., 2013). Furthermore, MAPK1 and related kinases have either reduced expression or phosphorylation in post-mortem brains of depressed individuals that committed suicide (Dwivedi et al., 2001, 2009).
It should be mentioned that there were limitations to this study. First, we only assessed the CP adjacent to the hippocampus. It is possible that the CP located in other regions of the brain may exhibit a different pattern of gene expression in MDD subjects. Moreover, we only assessed CP gene expression near the caudal hippocampus. This was due to the availability of high-quality sectioned material that also contained the CP. Interestingly, 40% of the depressed subjects included in this study committed suicide. It is possible that our results may not generalize to studies that use subjects with mild or moderate depression. Another limitation of the study was that we may have not been properly powered to validate some of the gene expression findings by qRT-PCR. To this end, we recognize that the findings from this study may not be particularly strong. Although we were limited by the number of high quality subjects available for this study, more subjects may be required to validate other transcripts changes, particularly those with lower fold-changes. It is important to note that we added new patients for the qRT-PCR study. Finally, the qRT-PCR primers did not always target to the same region as the microarray probe (e.g., MYH9). Thus, the specifics of the methodology helps explain why fewer transcripts than expected validated in this study.
The possibility that the extracellular matrix of the CP may be altered in depression has profound implications for identifying biomarkers in the CSF. Although many of the transcripts that were interrogated by qRT-PCR are not known to exist as proteins in the CSF, many of the collagens shown in Figure 3 are present as precursors or preproproteins (Bora et al., 2012). Since fibulin-2 (FBLN2) is a secreted protein that binds calcium and stabilizes the extracellular matrix, it would be interesting to determine whether FBLN2 may have mood altering effects and act as an antidepressant. Finally, since transcripts that interact with actin (i.e., MYH11, P2RY5) were also downregulated, there may be an altered cytoskeleton in MDD subjects.
In conclusion, the majority of the abundantly expressed transcripts in the CP are transcripts that code ribosomal proteins under normal physiological conditions. In subjects with MDD, several transcripts that interact with the extracellular matrix and cytoskeleton were decreased in the CP. Given the variety of functions performed by the CP and the identification of various transcripts linked to depression for the first time, the CP is a novel target for the development of therapeutics. With the majority of the transcripts downregulated, the structure of the CP is likely disrupted. Ways to boost the extracellular matrix or cytoskeleton should be beneficial in treating depression.
Author Contributions
Cortney A. Turner performed the study concept and design, laboratory procedures, data analysis, writing of the first draft of the manuscript and final revision of the manuscript. Robert C. Thompson performed data analysis, laboratory procedures and final revision of the manuscript. William E. Bunney supervised sample collection and final revision of the manuscript. Alan F. Schatzberg helped revise an earlier draft of the manuscript. Richard M. Myers assisted with an earlier version of the manuscript. Jack D. Barchas assisted with the final draft of the manuscript. Huda Akil supervised study design and manuscript revision. Stanley J. Watson supervised dissection of the material, study design and manuscript revision.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Claudio Caamano, Sharon Burke, Jennifer Fitzpatrick and Hui Li for the conduct of this work. This work was supported by NIMH Conte Center Grant P50 MH60398, NIDA P01 DA021633, The Office of Naval Research (ONR) Grants N00014-09-1-0598 and N00014-12-1-0366, the Pritzker Neuropsychiatric Disorders Research Consortium Fund LLC (http://www.pritzkerneuropsych.org), the Hope for Depression Research Foundation, NCRR Grant UL1RR024986, as well as a Rachel Upjohn Clinical Scholars Award to Cortney A. Turner. This work was also funded by the Pritzker Neuropsychiatric Disorders Research Consortium, which is supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. A shared intellectual property agreement exists between this philanthropic fund and the University of Michigan, Stanford University, the Weill Medical College of Cornell University, the University of California at Irvine, and the HudsonAlpha Institute for Biotechnology to encourage the development of appropriate findings for research and clinical applications.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fnhum.2014.00238/abstract
References
Amaral, D., and Insuasti, R. (eds.). (1990). Hippocampal Formation. San Diego, CA: Academic Press, Inc.
Armstrong, J. M., Krementsova, E., Michalek, A. J., Heaslip, A. T., Nelson, S. R., Trybus, K. M., et al. (2012). Full-length myosin Va exhibits altered gating during processive movement on actin. Proc. Natl. Acad. Sci. U.S.A. 109, E218–E224. doi: 10.1073/pnas.1109709109
Bachis, A., Mallei, A., Cruz, M. I., Wellstein, A., and Mocchetti, I. (2008). Chronic antidepressant treatments increase basic fibroblast growth factor and fibroblast growth factor-binding protein in neurons. Neuropharmacology 55, 1114–1120. doi: 10.1016/j.neuropharm.2008.07.014
Baghdassarian, D., Toru-Delbauffe, D., Gavaret, J. M., and Pierre, M. (1993). Effects of transforming growth factor-beta 1 on the extracellular matrix and cytoskeleton of cultured astrocytes. Glia 7, 193–202. doi: 10.1002/glia.440070302
Bernard, R., Kerman, I. A., Thompson, R. C., Jones, E. G., Bunney, W. E., Barchas, J. D., et al. (2011). Altered expression of glutamate signaling, growth factor, and glia genes in the locus coeruleus of patients with major depression. Mol. Psychiatry 16, 634–646. doi: 10.1038/mp.2010.44
Bora, A., Anderson, C., Bachani, M., Nath, A., and Cotter, R. J. (2012). Robust two-dimensional separation of intact proteins for bottom-up tandem mass spectrometry of the human CSF proteome. J. Proteome Res. 11, 3143–3149. doi: 10.1021/pr300057v
Calati, R., Crisafulli, C., Balestri, M., Serretti, A., Spina, E., Calabro, M., et al. (2013). Evaluation of the role of MAPK1 and CREB1 polymorphisms on treatment resistance, response and remission in mood disorder patients. Prog. Neuropsychopharmacol. Bol. Psychiatry 44, 271–278. doi: 10.1016/j.pnpbp.2013.03.005
Cameron, H. A., and McKay, R. D. (2001). Adult neurogenesis produces a large pool of new granule cells in the dentate gyrus. J. Comp. Neurol. 435, 406–417. doi: 10.1002/cne.1040
Cornford, E. M., Varesi, J. B., Hyman, S., Damian, R. T., and Raleigh, M. J. (1997). Mitochondrial content of choroid plexus epithelium. Exp. Brain Res. 116, 399–405. doi: 10.1007/PL00005768
de Vega, S., Iwamoto, T., and Yamada, Y. (2009). Fibulins: multiple roles in matrix structures and tissue functions. Cell. Mol. Life Sci. 66, 1890–1902. doi: 10.1007/s00018-009-8632-6
Ditzen, C., Tang, N., Jastorff, A. M., Teplytska, L., Yassouridis, A., Maccarrone, G., et al. (2011). Cerebrospinal fluid biomarkers for major depression confirm relevance of associated pathophysiology. Neuropsychopharmacology 37, 1013–1025. doi: 10.1038/npp.2011.285
Duman, R. S. (2004). Depression: a case of neuronal life and death? Biol. Psychiatry 56, 140–145. doi: 10.1016/j.biopsych.2004.02.033
Dwivedi, Y., Rizavi, H. S., Roberts, R. C., Conley, R. C., Tamminga, C. A., and Pandey, G. N. (2001). Reduced activation and expression of ERK1/2 MAP kinase in the post-mortem brain of depressed suicide subjects. J. Neurochem. 77, 916–928. doi: 10.1046/j.1471-4159.2001.00300.x
Dwivedi, Y., Rizavi, H. S., Zhang, H., Roberts, R. C., Conley, R. R., and Pandey, G. N. (2009). Aberrant extracellular signal-regulated kinase (ERK)1/2 signalling in suicide brain: role of ERK kinase 1 (MEK1). Int. J. Neuropsychopharmacol. 12, 1337–1354. doi: 10.1017/S1461145709990575
Fleming, C. E., Nunes, A. F., and Sousa, M. M. (2009). Transthyretin: more than meets the eye. Prog. Neurobiol. 89, 266–276. doi: 10.1016/j.pneurobio.2009.07.007
Gagelin, C., Pierre, M., Gavaret, J. M., and Toru-Delbauffe, D. (1995). Rapid TGF beta 1 effects on actin cytoskeleton of astrocytes: comparison with other factors and implications for cell motility. Glia 13, 283–293. doi: 10.1002/glia.440130405
Gazzin, S., Strazielle, N., Schmitt, C., Fevre-Montange, M., Ostrow, J. D., Tiribelli, C., et al. (2008). Differential expression of the multidrug resistance-related proteins ABCb1 and ABCc1 between blood-brain interfaces. J. Comp. Neurol. 510, 497–507. doi: 10.1002/cne.21808
Grimm, P. R., Irsik, D. L., Settles, D. C., Holtzclaw, J. D., and Sansom, S. C. (2009). Hypertension of Kcnmb1-/- is linked to deficient K secretion and aldosteronism. Proc. Natl. Acad. Sci. U.S.A. 106, 11800–11805. doi: 10.1073/pnas.0904635106
Gustafsson, E., Lindvall, O., and Kokaia, Z. (2003). Intraventricular infusion of TrkB-Fc fusion protein promotes ischemia-induced neurogenesis in adult rat dentate gyrus. Stroke 34, 2710–2715. doi: 10.1161/01.STR.0000096025.35225.36
Ishii, S., Noguchi, K., and Yanagida, K. (2009). Non-Edg family lysophosphatidic acid (LPA) receptors. Prostaglandins Other Lipid Mediat. 89, 57–65. doi: 10.1016/j.prostaglandins.2009.06.001
Johanson, C. E., Stopa, E. G., and McMillan, P. N. (2011). The blood-cerebrospinal fluid barrier: structure and functional significance. Methods Mol. Biol. 686, 101–131. doi: 10.1007/978-1-60761-938-3_4
Jones, E. G., Hendry, S. H., Liu, X. B., Hodgins, S., Potkin, S. G., and Tourtellotte, W. W. (1992). A method for fixation of previously fresh-frozen human adult and fetal brains that preserves histological quality and immunoreactivity. J. Neurosci. Methods 44, 133–144. doi: 10.1016/0165-0270(92)90006-Y
Kahl, K. G., Bens, S., Ziegler, K., Rudolf, S., Kordon, A., Dibbelt, L., et al. (2009). Angiogenic factors in patients with current major depressive disorder comorbid with borderline personality disorder. Psychoneuroendocrinology 34, 353–357. doi: 10.1016/j.psyneuen.2008.09.016
Kerman, I. A., Bernard, R., Bunney, W. E., Jones, E. G., Schatzberg, A. F., Myers, R. M., et al. (2012). Evidence for transcriptional factor dysregulation in the dorsal raphe nucleus of patients with major depressive disorder. Front. Neurosci. 6:135. doi: 10.3389/fnins.2012.00135
Kiec-Wilk, B., Stolarz-Skrzypek, K., Sliwa, A., Zdzienicka, A., and Kawecka-Jaszcz, K. (2010). Peripheral blood concentrations of TGFbeta1, IGF-1 and bFGF and remodelling of the left ventricle and blood vessels in hypertensive patients. Kardiol. Pol. 68, 996–1002.
Kratzer, I., Liddelow, S. A., Saunders, N. R., Dziegielewska, K. M., Strazielle, N., and Ghersi-Egea, J. F. (2013). Developmental changes in the transcriptome of the rat choroid plexus in relation to neuroprotection. Fluids Barriers CNS 10, 25. doi: 10.1186/2045-8118-10-25
Lanz, T. A., Bove, S. E., Pilsmaker, C. D., Mariga, A., Drummond, E. M., Cadelina, G. W., et al. (2012). Robust changes in expression of brain-derived neurotrophic factor (BDNF) mRNA and protein across the brain do not translate to detectable changes in BDNF levels in CSF or plasma. Biomarkers 17, 524–531. doi: 10.3109/1354750X.2012.694476
Li, J. Z., Vawter, M. P., Walsh, D. M., Tomita, H., Evans, S. J., Choudary, P. V., et al. (2004). Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum. Mol. Genet. 13, 609–616. doi: 10.1093/hmg/ddh065
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez-Figueroa, A. L., Norton, C. S., Lopez-Figueroa, M. O., Armellini-Dodel, D., Burke, S., Akil, H., et al. (2004). Serotonin 5-HT1A, 5-HT1B, and 5-HT2A receptor mRNA expression in subjects with major depression, bipolar disorder, and schizophrenia. Biol. Psychiatry 55, 225–233. doi: 10.1016/j.biopsych.2003.09.017
Mallei, A., Shi, B., and Mocchetti, I. (2002). Antidepressant treatments induce the expression of basic fibroblast growth factor in cortical and hippocampal neurons. Mol. Pharmacol. 61, 1017–1024. doi: 10.1124/mol.61.5.1017
Marques, F., Sousa, J. C., Coppola, G., Gao, F., Puga, R., Brentani, H., et al. (2011). Transcriptome signature of the adult mouse choroid plexus. Fluids Barriers CNS 8, 10. doi: 10.1186/2045-8118-8-10
Mercier, C., Masseguin, C., Roux, F., Gabrion, J., and Scherrmann, J. M. (2004). Expression of P-glycoprotein (ABCB1) and Mrp1 (ABCC1) in adult rat brain: focus on astrocytes. Brain Res. 1021, 32–40. doi: 10.1016/j.brainres.2004.06.034
Renard, M., Callewaert, B., Baetens, M., Campens, L., Macdermot, K., Fryns, J. P., et al. (2011). Novel MYH11 and ACTA2 mutations reveal a role for enhanced TGFbeta signaling in FTAAD. Int. J. Cardiol. 165, 314–321. doi: 10.1016/j.ijcard.2011.08.079
Roberts, L. M., Black, D. S., Raman, C., Woodford, K., Zhou, M., Haggerty, J. E., et al. (2008). Subcellular localization of transporters along the rat blood-brain barrier and blood-cerebral-spinal fluid barrier by in vivo biotinylation. Neuroscience 155, 423–438. doi: 10.1016/j.neuroscience.2008.06.015
Sathyanesan, M., Girgenti, M. J., Banasr, M., Stone, K., Bruce, C., Guilchicek, E., et al. (2012). A molecular characterization of the choroid plexus and stress-induced gene regulation. Transl. Psychiatry 2, e139. doi: 10.1038/tp.2012.64
Sequeira, A., Klempan, T., Canetti, L., ffrench-Mullen, J., Benkelfat, C., Rouleau, G. A., and Turecki, G. (2007). Patterns of gene expression in the limbic system of suicides with and without major depression. Mol. Psychiatry 12, 640–655. doi: 10.1038/sj.mp.4001969
Sequeira, A., Mamdani, F., Ernst, C., Vawter, M. P., Bunney, W. E., Lebel, V., et al. (2009). Global brain gene expression analysis links glutamatergic and GABAergic alterations to suicide and major depression. PLoS ONE 4:e6585. doi: 10.1371/journal.pone.0006585
Skipor, J., and Thiery, J. C. (2008). The choroid plexus–cerebrospinal fluid system: undervaluated pathway of neuroendocrine signaling into the brain. Acta Neurobiol. Exp. (Warsz). 68, 414–428.
Sullivan, G. M., Mann, J. J., Oquendo, M. A., Lo, E. S., Cooper, T. B., and Gorman, J. M. (2006). Low cerebrospinal fluid transthyretin levels in depression: correlations with suicidal ideation and low serotonin function. Biol. Psychiatry 60, 500–506. doi: 10.1016/j.biopsych.2005.11.022
Takebayashi, M., Hashimoto, R., Hisaoka, K., Tsuchioka, M., and Kunugi, H. (2010). Plasma levels of vascular endothelial growth factor and fibroblast growth factor 2 in patients with major depressive disorders. J. Neural Transm. 117, 1119–1122. doi: 10.1007/s00702-010-0452-1
Toro, B., Cox, N., Wilson, R. J., Garrido-Sanabria, E., Stefani, E., Toro, L., et al. (2006). KCNMB1 regulates surface expression of a voltage and Ca2+-activated K+ channel via endocytic trafficking signals. Neuroscience 142, 661–669. doi: 10.1016/j.neuroscience.2006.06.061
Turner, C. A., Clinton, S. M., Thompson, R. C., Watson, S. J. Jr., and Akil, H. (2011). Fibroblast growth factor-2 (FGF2) augmentation early in life alters hippocampal development and rescues the anxiety phenotype in vulnerable animals. Proc. Natl. Acad. Sci. U.S.A. 108, 8021–8025. doi: 10.1073/pnas.1103732108
Veeranna, Shetty, K. T., Takahashi, M., Grant, P., and Pant, H. C. (2000). Cdk5 and MAPK are associated with complexes of cytoskeletal proteins in rat brain. Brain Res. Mol. Brain Res. 76, 229–236. doi: 10.1016/S0169-328X(00)00003-6
Wolburg, H., and Paulus, W. (2010). Choroid plexus: biology and pathology. Acta Neuropathol. (Berl). 119, 75–88. doi: 10.1007/s00401-009-0627-8
Keywords: cytoskeleton, mRNA, hippocampus, depression, brain
Citation: Turner CA, Thompson RC, Bunney WE, Schatzberg AF, Barchas JD, Myers RM, Akil H and Watson SJ (2014) Altered choroid plexus gene expression in major depressive disorder. Front. Hum. Neurosci. 8:238. doi: 10.3389/fnhum.2014.00238
Received: 15 January 2014; Accepted: 02 April 2014;
Published online: 22 April 2014.
Edited by:
John J. Foxe, Albert Einstein College of Medicine, USAReviewed by:
Sara Morley-Fletcher, CNRS-University Lille, FranceWenyu Luo, University of California, San Francisco, USA
Michael Joseph McCarthy, Veterans Affairs San Diego Healthcare System, USA
Copyright © 2014 Turner, Thompson, Bunney, Schatzberg, Barchas, Myers, Akil and Watson. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cortney A. Turner, Molecular and Behavioral Neuroscience Institute, University of Michigan, 205 Zina Pitcher Place, Ann Arbor, MI 48109, USA e-mail: caturner@umich.edu
 Cortney A. Turner
Cortney A. Turner Robert C. Thompson
Robert C. Thompson William E. Bunney3
William E. Bunney3  Alan F. Schatzberg
Alan F. Schatzberg Richard M. Myers
Richard M. Myers Huda Akil
Huda Akil