Abstract
This article deals with the question of how self-organization in living organisms is realized. Self-organization may be observed in open systems that are out of equilibrium. Many disequilibria-conversion phenomena exist where free energy conversion occurs by spontaneously formed engines. However, how is self-organization realized in living entities? Living cells turn out to be self-organizing disequilibria-converting systems of a special kind. Disequilibrium conversion is realized in a typical way, through employing information specifying protein complexes acting as nano engines. The genetic code enables processing of information—derived from coding DNA—to produce these molecular machines. Hence, information is at the core of living systems. Two promising approaches to explaining living cells containing sequences carrying information are mentioned. Also discussed is the question of whether a second concept of self-organization—namely, the Kantian concept—applies.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Life and Explanation
Manifestations of life—we primarily discuss the unicellular life forms of prokaryotes—are usually easily recognizable. A characteristic of living entities is, for instance, their chemistry. They employ and build carbohydrates, lipids, proteins, and nucleic acids in characteristic ways. Any living cell includes a huge variety of molecular interactions (the interactome). Living cells are, among others, characterized by specific and selective far-from-equilibrium states. Descriptions of living entities often intermingle mechanical and biological terms.
Physicochemical phenomena demonstrate mechanical regularities, which enable causal explanation. Causal/mechanical and mechanistic explanation are renowned instances of causal explanation (De Regt 2011). According to Salmon’s causal/mechanical model, explaining a phenomenon consists of describing the causal processes and causal interactions that led up to that outcome (Salmon 1984). In mechanistic explanation, “phenomena are explained by describing the mechanisms that produce them” (De Regt 2011, p. 11). As defined by Machamer and colleagues (2000, p. 3), mechanisms are “entities and activities organized such that they are productive of regular changes from start or set-up to finish or termination conditions.” Causal/mechanical and mechanistic models both demonstrate that mechanical regularity enables causal explanation.
However, explanation of living entities encounters special problems. Because of maintaining low entropy, at first glance they seem to defy the second law of thermodynamics. A relevant definition and some explication of entropy is given by Balmer (2011, pp. 206, 207; italics in original): it is “a measure of the amount of molecular disorder within a system,” and “Entropy can only be produced (but not destroyed) within a system,” as the second law states. “The entropy of a system can be increased or decreased by entropy transport across the system boundary” (2011, p. 207; italics in original). Boltzmann pointed out that the second law depends essentially on probability theory. As a statistical law it constrains only the average run of events (Branscomb et al. 2017). Cannon’s term homeostasis (see Cannon 1929) distinguishes the steady state of living systems from physicochemical equilibrium. Thermodynamics shows that homeostasis, that is, biological stability, requires dynamic chemical activity (see the third section).
In 1944 Schrödinger ([1944]1992) published a—still discussed (see, e.g., Karaca 2019; Kauffman 2020)—booklet, titled “What is life?” In it he raises some questions. The first is: what is the source of the orderliness encountered in the unfolding of events in the life cycle of an organism? He supposes an “aperiodic crystal” containing a “code script,” calling it “the material carrier of life” ([1944]1992, pp. 5, 21). His second question is: how does living matter evade the decay to equilibrium? He indicates that its organization is maintained by extracting “order” from the environment. Schrödinger is focusing on the development of multicellular organisms whereas we focus primarily on single-celled organisms. Yet his questions about the source of orderliness and the evasion of decay to equilibrium are relevant for all living cells. Shortly after the publication of Schrödinger’s booklet, Bertalanffy (1950) made plausible that a necessary feature of all life forms is that a living entity is an open system. In the second half of the 20th century research on nonlinear dynamics, far-from-equilibrium open systems, and statistical physics has been consulted for better understanding life’s characteristics. Cells are the basic building blocks of all living systems. Cells are characterized by an order that cannot be achieved at thermodynamic equilibrium (cf. Brown and Sivak 2020). We have to look at systems far from equilibrium to grasp the generation of that order.
Self-Organization
In both animate and inanimate systems far from equilibrium, order can evolve by self-organization. “In [the] 1960s, Prigogine formalized the nature of dynamic self-organization as emergence of order in systems with […] an ability to dissipate the energy gradients effectively,” Demirel (2010, p. 958) indicates. Open systems far from equilibrium are characterized by irreversible, i.e., entropy-producing, processes. In a far-from-equilibrium condition nonlinear relationships prevail (Toffler 1984). Physicochemical processes may induce “the possibility of new types of instability, including the amplification of fluctuations breaking the initial spatial symmetry” (Prigogine and Stengers 1984, p. 148). Prigogine and Nicolis (1967, p. 3543) suggest “implications of such ‘self-organizing’ open systems for biogenetic processes.” However, with respect to self-organization conceptual questions can be raised. It is obvious that (physicochemical) self-organizing processes are important for living entities. However, in living cells particular self-organizing processes may be so entangled with genetic mechanisms—as we will see below—that the question can arise in what sense and to what extent these processes are self-organizing. We will give some attention to this question in the last section.
Branscomb et al. (2017, p. 5) state that, “many spontaneous disequilibria conversion phenomena exist that in a sense self-organize their own conversion mechanisms.” A conversion mechanism, they indicate, acts as an “engine,” lashing “the up-hill process to a more powerful one proceeding in its spontaneous, down-hill direction; in this way converting one disequilibrium into another” (2017, p. 1). Cottrell (1979) elaborates on this point, to which we will connect here. An “open system may ‘yield’ to the thermodynamic forces exerted upon it from its environment and, in so doing, thereby soak up free energy from that environment” (1979, p. 3). All systems extracting, concentrating, and storing free energy by acting on an energy flow—whether spontaneously formed, man-made, or biological—are engines because they generate working power from an energy input, Cottrell (1979) defines. He indicates that living entities are “composed of chemically unstable molecules, charged with free energy, delicately structured, yet able to survive for long periods and even to grow and multiply” (1979, p. 2). This raises the question: how do living things “capture free energy from the environment and maintain themselves in such statistically improbable structures” (1979, p. 2)? Cottrell remarks that in contrast to “engines formed spontaneously in an initially homogeneous medium, […] biological and man-made engines […] are not spontaneously created but are made or nucleated by pre-existent organized or structured systems” (1979, p. 6; italics in original).
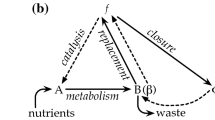
Eigen proposes a theory of self-organization in order to address the problem of interplay between cause and effect, and writes, “The present interplay of nucleic acids and proteins corresponds to a complex hierarchy of ‘closed loop’” (Eigen 1971, p. 467). Kauffman (1993, p. 295) considers life starting “as a minimally complex collection of peptide or RNA catalysts capable of achieving simultaneously collective reflexive catalysis of the set of polymers (hence replication of an autocatalytic set of polymer catalysts) and a coordinated web of metabolism.” The logic of this hypothesis fits well the observation that “dynamically coupled processes may lead to systems that acquire emergent properties that cannot be expected from individual processes” (Demirel 2010, p. 958).
In our discussions we employ the entropically-based model advanced by Branscomb et al. (2017). They indicate that in nonequilibrium thermodynamics disequilibria are pivotal. They assume that processes are driven by disequilibria—“gradients” between states that differ in their entropy content—and not by energy. As Branscomb and Russell (2013, p. 63) state: “A flux between these two entropy states, if not physically prevented, will arise spontaneously, driving the matter and/or energy from the state of low entropy to that of higher entropy.”
In accordance with Demirel’s representation of Prigogine’s formalization (see above), we define self-organization as emergence of order in systems with an ability to dissipate disequilibria effectively. Regarding living entities and self-organization many questions may be asked. However, in this article we confine ourselves to one question: how is self-organization realized in living cells?
Disequilibrium Conversion in Living Cells
What characterizes thermodynamics of living cells? Self-organization in living entities implies that these entities evade the decay to equilibrium. Living entities are thermodynamically open systems; they exchange matter and energy with their environment (Bertalanffy 1950). More precisely, all living systems exist in, and self-generate, a physical state that is far from thermodynamic equilibrium (cf. Branscomb and Russell 2018). To answer our research question we should know how living entities maintain a physical state that is far from equilibrium. It has been argued that this requires many instances of disequilibrium conversion, or—in other words—free energy conversion (FEC) (Branscomb and Russell 2013). But how are these conversions realized in living cells? “Mass action, quasi-equilibrium chemistry” is unable to perform this (Branscomb et al. 2017, p. 3), due to endergonic barriers, i.e., kinetically inhibited reactions (Barge et al. 2016). Branscomb et al. (2017, p. 5) summarize: “since mass action chemical reactions [i.e., all reactions that are possible near thermodynamic equilibrium], no matter how catalyzed, can only dissipate disequilibria—and cannot by themselves generate them, a fundamentally different mechanism is required, one which technically functions as an engine.”
Living entities necessarily generate and maintain “highly specific, dynamic, far-from-equilibrium states” (Branscomb et al. 2017, p. 5). Branscomb and Russell (2013, p. 63) indicate that, “the flow induced by a disequilibrium may be a compound process—i.e. composed of (at least) two processes, mechanistically linked so they function as a single process thermodynamically.” A molecular device couples—the manner of this coupling is indicated hereafter—an endergonic process (e.g., the reduction of CO2) to an exergonic process (e.g., the hydrolysis of ATP) “in order to drive the endergonic partner and thereby create further internal disequilibria.” This device is an engine for it accomplishes free energy conversion, and “all systems that carry out free energy conversion” are engines, and “all engines are FECs,” Branscomb and Russell (2013, p. 64) summarize Cottrell (1979). In line with the “entropically-based model” (Branscomb and Russell 2017, p. 40) in the present article (Gibb’s) free energy does not mean “physically an ‘energy’ (of any stripe),” but “just a dimensionless numerical quantity measuring how far from equilibrium the system currently is” (Branscomb and Russell 2013, p. 63).
Hoffman (2016, p. 3) defines a molecular machine as “a molecule or small molecular assembly that performs a function that locally increases free energy or performs work at the expense of chemical energy, while in the presence of significant thermal fluctuations.” He indicates that the general principles for molecular machines “touch on fundamental physical questions such as the meaning of entropy, free energy, and the second law of thermodynamics at the single molecule level” (2016, p. 2). Nano engines are macromolecular protein or protein-RNA complexes. The general operational requirements of these molecular engines differ greatly from macroscopic engines (Branscomb et al. 2017; cf. Purcell 1977). Important in this context is that, “the systems are inherently buffeted by unimaginably violent Brownian impacts” (2017, p. 6). These Brownian impacts induce significant thermal fluctuations, as we will see in the paragraph below.
Disequilibria conversions by nano engines appear to be an important part of the answer to Schrödinger’s second question (see above). Life needs “usable disequilibria.” “Disequilibria conversions of the kinds relevant to life cannot happen on their own,” as Branscomb and Russel (2018) indicate. An (endergonic or uphill) “driven” reaction, creating a specific disequilibrium, must be coupled by a case-specific “mechanochemical” macromolecular “engine” to an (exergonic or downhill) “driving” one, dissipating a greater disequilibrium. The engine operates as an escapement mechanism: the progress of the driving reaction depends on—is “gated by”—progress of the driven—endergonic!—reaction. The operating principle of a disequilibria-converting device is that, “the driving process is only allowed to complete in a given conversion cycle if the driven process has also completed in the same cycle” (Branscomb et al. 2017, p. 33; cf. Duval et al. 2013). However, the question arises: do instances of the endergonic direction of the driven reaction occur? The answer is: yes, Brownian impacts induce both “backward” and “forward” reactions, although not in equal amount, as the second law reminds us. The case-specific engine, the escapement mechanism, is a stochastic device that uses probable “forward” thermal fluctuations in one (driving) process to select improbable “reverse” fluctuations in another (driven) process (Branscomb and Russell 2018). The two reactions are coupled together “into a single—and spontaneous—thermodynamic process” (Branscomb and Russell 2013, p. 62). Notice that these self-organizing processes in living entities require the preceding presence of specific protein complexes, and therefore, genetic mechanisms. As Newman and Comper (1990, p. 1) indicate, genetic mechanisms are “highly evolved, machine-like, biomolecular processes.”
Branscomb et al. (2017, p. 33) state that, “the protein complex-based engines powering life by interconverting chemical disequilibria achieve their escapement functions by moving through a corresponding series of enzyme forms, or ‘alters’ [i.e., allosteric changes] each having its own set of binding and catalytic specificities.” To construct protein complexes acting as machines, cells necessitate polypeptides with specific sequences of amino acids. Proteins are named “dynamic actors” (Bu et al. 2011, p. 163). This applies to protein complexes acting as disequilibria-converting nano engines, but also for enzymes, which are highly selective and specific catalysts, and for dynamic structural elements (cf. Branscomb and Russell 2018). Living cells employ highly selective and specific molecular machines to maintain homeostasis. Hoffman (2016, p. 17) indicates that, “chemically induced conformational changes, i.e., allostery,” are “absolutely essential for real molecular machines.” Cells employ proteins with an arrangement of amino acids capable of driving selectively specific chemical reactions that constitute the metabolism. A cell needs more than its physicochemical equipment to accomplish the formation of necessary, functional proteins. It needs a form of steering polypeptide synthesis in order to obtain the required protein complexes for cell functioning, including engines for the conversion of disequilibria as explained above. Such a form of steering is provided by information in coding DNA/RNA, as we will see in the next section.
Cells Employ Information for Constructing Molecular Machines
The Genetic Code and Protein Biosynthesis
Cells employ specified proteins for disequilibrium conversions. To specify the proteins that will be synthesized, a cell employs parts of its DNA. DNA is a polymer consisting of four different nucleotides. A nucleotide contains a sugar (deoxyribose), a phosphate group, and an organic base. The nucleotides differ in their base molecule. The four base molecules contained in DNA are guanine (G), adenine (A), cytosine (C), and thymine (T). A DNA molecule is a double helix of two complementary strands of nucleotides, which means the bases are complementary in this double helix structure: C in one strand is always opposite G in the other strand and A opposite to T. In cell division, the DNA is replicated—a copying process—resulting in two molecules, both identical to the original molecule. In metabolic functioning, parts of the DNA become transcribed into functionally different kinds of RNA. RNA differs from DNA in being usually single-stranded, in being constructed with a different sugar (ribose) in the four nucleotides, and in substituting uracil (U) for thymine. For making proteins, specific parts of DNA—named protein-coding regions (or in short form, coding regions)—are transcribed into RNA, called messenger RNA (mRNA), which at the ribosomes is translated into polypeptides by a nearly universal code—named the genetic code.
In the genetic code, a sequence of three nucleotides/bases in DNA forms a so-called codon, coding for one out of the 20 possible amino acids that naturally occur in proteins. In protein synthesis a codon has a sign function. In transcription, DNA functions as a template for one complementary RNA strand. Translation of the nucleotide sequence of mRNA into an amino acid sequence in a protein occurs at the ribosomes (consisting of rRNA and proteins). In this process transfer-RNA (tRNA) molecules are involved, functioning as adaptors which carry the amino acids, and also specific enzymes, to combine a codon with the specific amino acid. Other parts of DNA are translated into RNA with functions in regulation or immunity.
Statistical Information
In discussing information contained within sequences, often Shannon entropy is introduced as an important concept. This prompts us to consider that concept. DNA contains signs that are segregated (they do not blend), linear (they are in a sequence), and digital (their functioning as a sign is not analogous, but like a digit) (cf. Yockey 2005). DNA is made up of two complementary (long) strands containing four different bases (see previous subsection)—functioning informationally. Shannon (1948) has investigated the capacity of information channels. His work shows that a string of signs—as the codons in DNA do form—may contain information (or: data). In information theory Shannon entropy is a measure of choice or uncertainty (Shannon 1948). Regarding the statistics of information Shannon entropy is a measure of information (Weaver [1949]1964; Yockey 2005). As Shannon himself mentioned, he disregarded the semantics of information. The word information in his mathematical theory of communication, i.e., in information theory, must not be confused with meaning: it is used in a special sense (Weaver [1949]1964). However, to value the specific information in sequences of coding regions we need precisely to pay attention to the meaning that this information has in the context of the living cell, including its functioning genetic code.
Semiosis
Semiosis is the production, action, and interpretation of signs (Queiroz et al. 2011). The study of sign systems is called semiotics. A sign is a representation of something other than itself. Since Peirce’s famous “New List” (Peirce [1867]1868) semioticians distinguish (cf., e.g., Jakobson 1974) iconic, indexical, and symbolic signs. Icons represent objects by likeness—e.g., a portrait represents a person—whereas indices point to objects with which they are physically related (a relation by “a correspondence in fact,” as Peirce ([1867]1868, p. 294) formulates)—e.g., smoke is an index of fire. Deacon (2012, p. 11) points out that a symbolic sign relationship “involves a conventional sign type that is additionally conventionally-mediated in the way it represents.” In this article we make no claims about the typology of signs, i.e., we do not discuss which kind of signs—iconic, indexical, or symbolic—are involved.
Do we find sign processes in living cells? In protein-coding DNA (or in short form: coding DNA), a specific codon, that is, a triplet of bases, indicates a specific amino acid (see above). Living cells appear to employ sequences of nucleotides as natural signs. Consequently, we do find sign processes in living cells. The study of sign processes in living systems is named biosemiotics. If confined to processes at the cellular and subcellular level, it has also been named cytosemiotics (e.g., Sebeok 2001). What are the semiotic characteristics of protein biosynthesis in living cells? In the remainder of this article, we will explicate some characteristics and point a direction for further research.
Information in Sequences
Watson and Crick (1953) discovered in DNA the informational functioning of patterns of bases. They define “the code which carries the genetic information” as “the precise sequence of the bases” (1953, p. 965). The word code has been used in two different ways: (1) DNA sequences can be said to affect processes by containing a code—this is code according to Watson and Crick (1953; cf. Trifonov 2011). (2) DNA sequences can be said to affect processes by a set of rules, being a code, that establishes a correspondence (or mapping) between two independent worlds—this is code according to Barbieri (2003; cf. Yockey 2005). In this article we name the first definition of code, viz. the precise sequence of the bases in protein-coding DNA—information (not “a code”). The second definition of code—a mapping between two independent worlds—accords with the genetic code. These two phenomena are complementary. Both information and the genetic code are necessary for the self-organization in cells described above.
The genetic code enables the processing of information. Through an invariant genetic code the transcription and translation of a nucleotide sequence in DNA results in a specific polypeptide. At the same time we know there are many other “sequence codes” in living cells, as Trifonov (2011) calls them. A sequence code has been defined by Trifonov as “any pattern or bias in the sequence which corresponds to one or another specific biological (biomolecular) function or interaction” (2011, p. 2).
A DNA sequence may convey a maximum amount of information if the four organic bases in DNA have a physiochemically equal probability of forming any particular element of a series (Polanyi 1967, 1968). For DNA this has appeared to be the case completely or almost completely. Subsequently, arbitrary relations—not dictated by physical necessities—are realized between DNA sequences and cell processes by the interference of adaptors (Barbieri 2015).
Information in Protein-coding DNA/RNA
Specific sequences of bases in nucleic acids contain—in the context of a certain cell—specific information. We define information in a living cell as a pattern of signs, which—by a decoding process in that cell—delivers function to the cell. With regard to protein-coding DNA, specifying means the pattern—in this case a linear order—is coding for functionally appropriate proteins. Often but not always this means the protein is appropriate to function as an active enzyme, whether or not acting as an engine or as a part thereof. Specification is realized in the context of a living cell. Therefore, information can only have this meaning for a living cell. According to the above definition, a protein does not carry information. Bases in protein-coding DNA, however, function in the cell as signs as they stand for something else according to physicochemical arbitrary rules (see above). This justifies speaking about an informational process. In making proteins, the cell employs base sequences of DNA to determine amino acid sequences of proteins. In other words, protein-coding DNA provides “the specification of the amino acid sequence of the protein” (Crick 1958, p. 144). How can this phenomenon be explained? Does biological explanation coincide with physicochemical explanation? In a physicochemical way, information is imparted by DNA in the life process, but is the information itself a physicochemical phenomenon? Scientists have been struggling with these questions for a long time. In particular, Niels Bohr (1885–1962) and Michael Polanyi (1891–1976) have published helpful considerations.
In his 1932 lecture Light and life, Bohr (1933) has pointed to the two complementary descriptions of light—viz. light as a wave phenomenon and light as particles—that physicists need to explain the different light phenomena. He proposes an analogy with the attitude that biologists more or less intuitively have adopted towards the aspects of life. Bohr emphasizes the simplicity of the phenomenon of light compared to the complexity of life. In a 1936 lecture, Bohr argues that developments in nuclear physics require us to replace in the case of light the ideal of causality with complementarity. In this lecture (Bohr 1937) he points to the possibility that, “essential features of living organisms [….] are laws of a nature which stands in a complementary relationship to those with which we are concerned in physics and chemistry” (1937, p. 296). In the same lecture he also indicates that,
this situation […] implies no limitation whatever in the application to biology of the physicochemical methods of description and investigation; in fact, the appropriate use of such methods—just as even in atomic physics all our experiences must rest upon experimental arrangements classically described—remains our sole and inexhaustible source of information about biological phenomena. (1937, p. 297)
Polanyi (1968, p. 1308) indicates that “the organism is shown to be, like a machine, a system which works according to two different principles: its structure serves as a boundary condition harnessing the physical-chemical processes by which its organs perform their functions.” He argues (Polanyi 1967, p. 65) that, “the mere existence of machinelike functions in living beings proves that life cannot be explained in terms of physics and chemistry.” Irreducible higher principles, that is, principles that are additional to the laws of physics and chemistry, characterize living entities (Polanyi 1968). We may also say that the living cell is a system that works according to two different principles: its structure serves as a boundary condition harnessing the physicochemical processes by which its parts—among them nano engines—perform their function. The living cell can aptly be named “a system under dual control” (cf. Polanyi 1968), viz. physicochemical and biological. Information—although being materially embodied—is itself not a physicochemical phenomenon, but a higher-level phenomenon imposing boundary conditions on the laws of physics (cf. Polanyi 1967; 1968).
In this article we do not discuss the role of information in living cells in general, but we highlight merely the specification given by information in protein-coding regions of the genome and transcripts thereof. In light of contemporary knowledge of cell and molecular biology, the presence of this information appears a necessary condition for living cells, as we know them, to maintain, for instance, the indispensable free energy conversion (FEC) processes. Protein-coding sequences have a double role in the cell because in addition to being informational these sequences are a physical constituent of the cell. Due to the latter the information is available for the cell to transcribe and to translate (see above). The production of biologically functional peptides requires (among other things) both the physicochemical equipment and specific information.
Protein-coding DNA/RNA and Function
The genetic code guarantees consistency in translation. Without consistent “translation”—matching with the specific information in the protein-coding DNA/RNA, i.e., changing the code necessitates changing the sequence of bases in the DNA/RNA sequences—the information cannot be meaningful, i.e., it cannot deliver a specific protein performing a function (cf. Gerstein et al. 2007) in the cell.
Moss (2003) differentiates two concepts of “the gene,” Gene-P and Gene-D. Gene-P is defined strictly on the basis of its instrumental utility in predicting a phenotypic outcome, and Gene-D is defined by its molecular sequence. Gene-D “provides possible templates for RNA and protein synthesis but has in itself no determinate relationship to organismal phenotypes” (2003, p. xiv). This description of Gene-D suits well with the protein-coding regions, coding for parts of nano engines, the subject of our study. The production of molecular engines by a cell requires—next to physical equipment—information specifying for functional proteins. Gerstein et al. (2007, p. 679) note: “DNA sequences determine the sequences of functional molecules” (cf. Crick 1958). Nano engines in the cell are part of the boundary condition of the cell (see previous subsection). For manufacturing them, the cell employs “specific biochemical information” (Nirenberg 1963, p. 80). Of course, for an extended evaluation of the role of information in living entities, it will be necessary to value other factors in determining functional proteins, as indicated by, for instance, posttranslational modification and alternative splicing (cf., e.g., Gerstein et al. 2007). Then, the role of gene regulation has to be taken into consideration in explaining cell functioning. However, that is not the subject of this article. Here, we merely indicate that cells employ information obtained from nucleic acids for making proteins acting as machines, among which are engines. We discuss the definition of gene-D only from this perspective.
Information at the Core of Living Entities
We agree with the designation self-organizing dissipative systems for living cells (Branscomb and Russell 2013; cf. Demirel 2010), at the same time emphasizing that this is not self-organization by spontaneously formed engines (see above). Branscomb and Russell (2013, p.70) designate living systems to be “organismically” “autocatalytic ‘self-organizing dissipative systems.’” However, more can be said about the mechanisms involved. Matter and energy in the dissipative system of the living cell are selectively controlled by the-cell-in-its-environment employing specific information stored in protein-coding DNA. For its characteristic self-organization, a living cell—whether unicellular or as part of a multicellular organism, whether prokaryote or eukaryote—necessitates functional protein complexes, acting as engines. Here, cellular self-organizing dissipative systems are realized through genetic mechanisms, as has been demonstrated above. This is not to deny that in living cells generic mechanisms—i.e., “those physical processes that are broadly applicable to living and non-living systems” (Newman and Comper 1990, p. 1)—are important, self-organizing processes in the forming of proteins included. We merely underscore that for its typical self-organization, a cell depends inter alia on its genetic code, and the information in precise sequences of bases in its coding DNA as a “template” for the synthesis of specific peptides.
Regarding Schrödinger’s first question (see first section), our results—which are limited to coding sequences—support the designation that an “aperiodic crystal” contains a “code-script” as a template for proteins. Some of these proteins are used for engines involved in disequilibria conversions (see third section). This leads to an answer to Schrödinger’s second question. The “code-script” enables “order-from-order” (cf. Kauffman 2020). However, Schrödinger’s designation “material carrier of life” overestimates the genetic material an sich, and underestimates the role of the rest of the cell (epigenetic chromosome modifications included) and the environment of the cell. Moss (2003, p. 61) aptly points out that there are “other sources” of order in the cell too. In addition, we would underline that the coding sequences only appear ordered in the context of the whole of the living cell (see above). Therefore, the answer to Schrödinger’s first question has to be understood as cell centered, not gene centered (cf. Harold 2001). Moss (2003, p. 77) points out that the relationship of each of three epigenetic inheritance systems—i.e., organizational structure, steady-state dynamics, and chromosome marking—to the genome (and to each other) is that of codependence and causal reciprocity. Additionally, we point out the indispensable role of information in coding DNA/RNA for the “correct synthesis” (Moss 2003, p. 81) of functional proteins.
Signaling the importance of specific information for cell functioning (cf. also Küppers 2010; Keller 2009) leads us to consider the hypothesis that a living cell, in its entirety, is both a physical system and “complementarily”—sensu Bohr—a semiotic system.
“Big questions” remain. How did the newness of biological entities emerge? One can ask how the physical basis of self-organization and the semiotic nature of genes became integrated in the whole of the living entity. How has the functional relationship arisen between cellular processes and specific sequences of bases in DNA? How did a system that develops merely through physical regularity and chance evolve into a system that is under dual control? In other words, as Pattee (1969) formulates: “How does a molecule become a message?” This question can be approached from primarily a physical perspective (see, for example, Newman et al. 2006). Here, we pass by this perspective and call attention to a semiotic perspective. We especially notice two promising semiotic explanatory approaches.
Explaining Information
A Mechanistic Approach
Eigen (1971, p. 468) states that, “the information resulting from evolution is a ‘valued’ information and the number of bits will not tell us too much about its functional significance.” Our interest is with the origination of the first biological information. According to Lehn (2013, p. 2838), “there must have been a purely chemical evolution that progressively led to the threshold of life.” Yockey (2005, p. 5; cf. Monod 1970) questions this approach, stating that, “if genetical processes were just complicated biochemistry, the laws of mass action and thermodynamics would govern the placement of amino acids in the protein sequences.” According to his understanding, “the process of the origin of life is possible but unknowable” (Yockey 2005, p. 173). However, according to physicochemical explanatory models, information in living cells must first have arisen by selective self-organization and evolution of macromolecules (cf. Küppers 1990; Kauffman 1993).
Code biology,Footnote 1 sometimes called code-based biosemiotics (e.g., Barbieri 2008b), is a mechanistic approach to biology giving centrality to the origination of natural conventions, based on coding. Barbieri (2015, 2016)—fostering “code biology”—remarks that biological processes cannot be reduced to physical quantities, because physical quantities only completely describe spontaneous systems, i.e., here, systems not manufactured by molecular machines. He elaborates “the idea that life is artifact making” (Barbieri 2015, p. 19f; emphasis in original), arguing that, “life is made of objects that are manufactured by molecular machines.” Barbieri (2015, p. 16; cf. 2019) defines mechanism as “scientific modelling” and distinguishes it from physicalism and (explanatory) reductionism. In discussing extensions of the Modern Synthesis, Barbieri (2015, p. 178) indicates that, “that there are two distinct types of evolutionary change: evolution by natural selection, based on copying, and evolution by natural conventions, based on coding.” Barbieri (2008a) discerns many codes in nature (not all related to DNA-sequences), calling them organic codes, and discusses the genetic code and the signal transduction codes at length (e.g., Barbieri 2015). He distances himself from “the endorsement of non-mechanism, or qualitative organicism” of Peircean biosemiotics (Barbieri 2015, p. 161; cf. 2003), which we will discuss below. According to him, cellular signs depend strictly on the codemaker—i.e., the translation apparatus. No interpretation is involved—as in sign-based biosemiotics (the subject of the next subsection) is assumed.
A Qualitative Organicist Approach
The writings of the philosopher and scientist Charles Sanders Peirce (1839–1914) have inspired many biologists to study living entities from the point of view of semiotics (cf. Emmeche and Kull 2011). However, as Emmeche (2011, p. 98) indicates: “Biosemiotics typically does not use Peirce’s broad ontological notion of life, but construes a notion of life derived from contemporary biology, […] life as organic sign-interpreting systems. But biosemiotics entails a thesis of the reality of ideal objects.” Queiroz et al. (2011, p. 91) consider life “not just from the perspectives of physics and chemistry, but also from a view of living systems that stresses the role of signs conveyed and interpreted by other signs in a variety of ways, including by means of molecules.” Emmeche et al. (2010, p. 635) claim “that the notion of information and other related ideas grasp some fundamental features of biological systems and processes that might be otherwise neglected.”
Emmeche (2004, p. 207f) describes mainstream organicism as “claiming biology to be an autonomous science, the organism to be ontologically real and irreducible to mere chemistry, though constituted by no other qualities than the emergent material properties of highly self-organized matter.” He continues, “the other option represents a more qualitative view on living beings; it emphasizes not only the ontological reality of biological higher level properties or entities […] but also the existence of phenomenological or qualitative aspects of at least some higher level properties,” naming this other option qualitative organicism. The approach of the qualitative organicist semioticians in biology—often shortly called biosemioticsFootnote 2—could also aptly be named sign-based biology (cf. Hoffmeyer 2010; Emmeche and Kull 2011).
For Peirce, any sign is something that stands for something else (its object) in such a way that it ends up producing a third relational entity (an interpretant), which is the effect a sign produces on an interpreter. In the context of biosemiotics, an interpreter is a biosystem such as a cell or an organism. The meaning of a sign can be accessed only through that sign’s effect (interpretant) upon some interpreting system, such as a biosystem. Granted that the cell is an interpreting system, where in the cell can we find the signs that harbor a message for that system? DNA has proven to contain signs for the cell system, as we saw above. Understandably, interpretation is a pivotal concept in (sign-based) biosemiotics. Kull (2011, p. 118) states: “An object is semiotic if it is in interpretation. Interpretation, according to the contemporary biosemiotic view, starts with the very process of life.” However, within Peircean-based biosemiotics bolder metaphysical claims are also made. Deely (2014) examines the proposition that semiosis preexisted living entities.
Perspective
We acknowledge a semiotic approach in (mechanistic) code biology and (organicist) biosemiotics. Both code biology and biosemiotics are promising tools in research on information. Barbieri elaborates a mechanistic approach without limiting himself to physicalist explanations. He defines (Barbieri 2016) (organic) information and (organic) meaning in empirical scientific terms. Peircean biosemioticians give attention to semiotic peculiarities of manifestations of life, and to biophysical and biochemical processes as part of an interpreting system. Is an integrated synthesis possible between these approaches, and how would it look? In a forthcoming article we will explore synchronic—i.e., conceptual— and diachronic—i.e., evolutionary—aspects of information. In that paper we plan to elaborate a semiotic approach.
Discussion and Conclusion
Branscomb and Russell (2018, p. 3) aptly explicate that, “an elaborate economy of disequilibria conversions” belongs to “the inherent nature of life.” We add that these disequilibria conversions in living cells (see the third section) depend on genetic mechanisms. In all observed living cells specificity and selectivity characterizing free energy conversion depend on information stored in protein-coding DNA/RNA. Specific information seems necessary for creating characteristic disequilibria. The observed manifestations of life suggest that the presence of protein-coding DNA/RNA containing information specifying functional proteins is a necessary condition for living entities. We conclude that the typical self-organization in living cells is only realized through employing information by the cell. At the same time, we endorse that living cells are self-organizing systems in the sense of the definition above (see the second section): order emerges within the cell, by the cell system dissipating disequilibria effectively.
Conceptual questions can arise regarding living cells and self-organization (second section). At the end of the 18th century, the philosopher Immanuel Kant ([1790]1908, p. 374) conceptualized a living entity as a “natural end” (Naturzweck), i.e., an “organized and self-organizing being” (organisiertes und sich selbst organisierendes Wesen). “Natural end,”—i.e., internal purposive organization—is a merely heuristic concept. It is not meant as an explanation, but to identify the object of biological research. Kant’s approach appeared productive in 19th-century biology (Beekman and Jochemsen 2022). His usage of the term “self-organization” may even be called “highly prescient” in light of later developments in thermodynamics and dynamics (Newman 2022, p. 215). Molecular biology has “molecularized” Kant’s concept of natural end (cf. Roth 2014) by making plausible that the concept of natural end applies at the cellular level, on which it finds expression in specific order at the molecular level. Therefore, we suggest that the title of our article also applies if “self-organization” is taken in Kant’s sense of the term (cf. Ginsborg 2001; Keller 2007). Whereas Prigogine-Demirel’s definition concerns physical self-organization—for instance in living entities, Kant discusses biological self-organization by living entities. We point out that both are found in any living cell.
Above we discuss two questions Schrödinger poses. But Schrödinger ([1944]1992, p. 76) also poses a third question: “Is life based on the laws of physics?” In his answer he refers to quantum theory. However, he just comments on “order-from-order,” and leaves unexplained the origination of the information employed in the cell. He also discusses “new laws to be expected in the organism” ([1944]1992, p. 76). Ever since Schrödinger’s booklet, no new physical laws have been formulated explaining manifestation of life. Walker and Davies (2016, p. 10) state that current physical theory—current paradigms—is not adequate to explain the “hard problem of life,” i.e., “that life seems distinct from other physical systems in how information affects the world.” We suggest that informing by semiosis is a necessary condition for the manifestation of life for it imposes essential boundary conditions on physical laws in living systems (the fourth section). We hypothesize that a living cell, in its entirety, is both a physical system and “complementarily”—sensu Bohr—a semiotic system. However, living after the mentioned discovery by Watson and Crick, we disagree with Bohr that physicochemical methods are the sole source of information about biological phenomena. We suggest that semiotics has to be part of biological method.
Data Availability
Not applicable.
Code Availability
Not applicable.
Notes
Some practitioners of this approach are united in the International Society of Code Biology. See www.codebiology.org.
Some practitioners of this approach are united in the International Society for Biosemiotic Studies. See www.biosemiotics.org.
References
Balmer RT (2011) Modern engineering thermodynamics. Elsevier, Amsterdam
Barbieri M (2003) The organic codes: an introduction to semantic biology. Cambridge University Press, Cambridge
Barbieri M (2008a) Life is semiosis: the biosemiotic view of nature. Cosmos and History 4(1–2):29–51
Barbieri M (2008b) Biosemiotics: a new understanding of life. Naturwissenschaften 95:577–599. https://doi.org/10.1007/s00114-008-0368-x
Barbieri M (2015) Code biology: a new science of life. Springer, Cham
Barbieri M (2016) What is information? Phil Trans R Soc A 374:20150060. https://doi.org/10.1098/rsta.2015.0060
Barbieri M (2019) A general model on the origin of biological codes. BioSystems 181:11–19. https://doi.org/10.1016/j.biosystems.2019.04.010
Barge LM, Branscomb E, Brucato JR, Cardoso SSS, Cartwright JHE, Danielache SO et al (2016) Thermodynamics, disequilibrium, evolution: far-from-equilibrium geological and chemical considerations for origin-of-life research. Orig Life Evol Biosph 47(1) (2017):39–56
Beekman W, Jochemsen H (2022) The Kantian account of mechanical explanation of natural ends in eighteenth and nineteenth century biology. Hist Philos Life Sci 44:10. https://doi.org/10.1007/s40656-022-00484-0
von Bertalanffy L (1950) The theory of open systems in physics and biology. Science 111:23–29. https://doi.org/10.1126/science.111.2872.23
Bohr N (1933) Light and life. Nature 133:421–423
Bohr N (1937) Causality and complementarity. Philos Sci 43:289–298
Branscomb E, Biancalani T, Goldenfeld N, Russell M (2017) Escapement mechanisms and the conversion of disequilibria; the engines of creation. Phys Rep 677:1–60. https://doi.org/10.1016/j.physrep.2017.02.001
Branscomb E, Russell MJ (2013) Turnstiles and bifurcators: the disequilibrium converting engines that put metabolism on the road. Biochim et Biophys Acta - Bioenergetics 1827:62–78. https://doi.org/10.1016/j.bbabio.2012.10.003
Branscomb E, Russell MJ (2018) Frankenstein or a submarine alkaline vent: who is responsible for abiogenesis? Part 1: What is life—that it might create itself? BioEssays 40.7: 8 pp. https://doi.org/10.1002/bies.201700179
Brown AI, Sivak DA (2020) Theory of nonequilibrium free energy transduction by molecular machines. Chem Rev 1201:434–459. https://doi.org/10.1021/acs.chemrev.9b00254
Bu Z, Callaway DJE (2011) Proteins move! Protein dynamics and long-range allostery in cell signaling. Adv Protein Chem Struct Biology 83. https://doi.org/10.1016/B978-0-12-381262-9.00005-7
Cannon WB (1929) Organization for physiological homeostasis. Physiol Rev 93:399–431
Cottrell A (1979) The natural philosophy of engines. Contemp Phys 201:1–10. https://doi.org/10.1080/00107517908227799
Crick FHC (1958) On protein synthesis. Symp. Soc. Exp. Biol 12: 138–163
Deacon TW (2012) Beyond the symbolic species. In: Stjernfelt F, Schilhab T, Deacon TW (eds) The symbolic species evolved. Springer, Dordrecht, pp 9–38. https://link.springer.com/book/10.1007/978-94-007-2336-8
Deely J (2014) Physiosemiosis as an influence of signs. How would semiosis precede life? Chin Semiotic Stud 103:375–407. https://doi.org/10.1515/css-2014-0033
Demirel Y (2010) Nonequilibrium thermodynamics modeling of coupled biochemical cycles in living cells. J Nonnewton Fluid Mech 165:953–972. https://doi.org/10.1016/j.jnnfm.2010.02.006
Duval S, Danyal K, Shaw S, Lytle AK, Dean DR, Hoffman BM et al (2013) Electron transfer precedes ATP hydrolysis during nitrogenase catalysis. PNAS USA 11041:16414–16419. https://doi.org/10.1073/pnas.1311218110
Eigen M (1971) Selforganization of matter and the evolution of biological macromolecules. Die Naturwiss 5810:465–523
Emmeche C (2004) Organicism and qualitative aspects of self-organization. Revue Int de philosophie 228:205–217. https://www.cairn.info/revue-internationale-de-philosophie-2004-2-page-205.ht
Emmeche C (2011) Organism and body: the semiotics of emergent levels of life. In: Emmeche C, Kull K (eds) Towards a semiotic biology: life is the action of signs. Imperial College Press, London, pp 91–111
Emmeche C, Kull K (eds) (2011) Towards a semiotic biology: life is the action of signs. Imperial College Press, London
Emmeche C, Queiroz J, El-Hani C (2010) Information and semiosis in living systems: a semiotic approach. In: Favareau D (ed) Essential readings in biosemiotics: anthology and commentary. Springer, Dordrecht, pp 629–656. https://doi.org/10.1007/978-1-4020-9650-1_20
Gerstein MB, Bruce C, Rozowsky JS, Zheng D, Du J, Korbel JO et al (2007) What is a gene, post-ENCODE? History and updated definition. http://www.genome.org/cgi/doi/10.1101/gr.6339607. Accessed 31 Mar 2022
Ginsborg H (2001) Kant on understanding organisms as natural purposes. In: Watkins E (ed) Kant and the sciences. Oxford University Press, New York, pp 230–258
Harold FM (2001) The way of the cell: molecules, organisms and the order of life. Oxford University Press, New York
Hoffman PM (2016) How molecular motors extract order from chaos. Rep Prog Phys 79(3). https://doi.org/10.1088/0034-4885/79/3/032601
Hoffmeyer J (2010) God and the world of signs: semiotics and the emergence of life. Zygon 45(2):367–390
Jakobson R (1974) Form und Sinn: Sprachwissenschaftliche Betrachtungen. Wilhelm Fink Verlag, München
Kant I [1790](1908) Kritik der Urteilskraft. Akademie Ausgabe Kants gesammelte Schriften, vol 5. Reimer, Berlin
Karaca C (2019) Relational basis of the organism’s self-organization: a philosophical discussion. Dissertation, University of Exeter
Kauffman SA (1993) The origins of order: self-organization and selection in evolution. Oxford University Press, New York
Kauffman SA (2020) Answering Schrödinger’s “What is life? Entropy 22(8):10. https://doi.org/10.3390/e22080815
Keller EF (2007) Disappearance of function from ‘self-organizing systems’. In: Boogerd FC, Bruggeman F, Hofmeyr J-H, Westerhoff HV (eds) Systems biology: philosophical foundations. Elsevier, Amsterdam, pp 303–317
Keller EF (2009) Rethinking the meaning of biological information. Biol Theory 4(2):159–166. https://doi-org.ezproxy.library.wur.nl/https://doi.org/10.1162/biot.2009.4.2.159
Kull K (2011) Life is many, and sign is essentially plural: on the methodology of biosemiotics. In: Emmeche C, Kull K (eds) Towards a semiotic biology: life is the action of signs. Imperial College Press, London, pp 113–129
Küppers B-O (1990) Information and the origin of life. MIT Press, Cambridge
Küppers B-O (2010) Information and communication in living matter. In: Davies P, Gregersen NH (eds) Information and the nature of reality: from physics to metaphysics. Cambridge University Press, Cambridge, pp 170–184
Lehn J-M (2013) Perspectives in chemistry – steps towards complex matter. Angewandte Chemie Int Ed 52:2836–2850. https://doi.org/10.1002/anie.201208397
Machamer P, Darden L, Craver CF (2000) Thinking about mechanisms. Philos Sci 67(1):1–25
Monod J (1970) Le hazard et la nécessité: essai sur la philosophie naturelle de la biologie moderne. Éditions du Seuil, Paris
Moss L (2003) What genes can’t do. MIT Press, Cambridge
Newman SA (2022) Self-organization in embryonic development: myth and reality. In: Malassé AD (ed) Self-organization as a new paradigm in evolutionary biology: from theory to applied cases in the tree of life. Springer, Cham, pp 195–222
Newman SA, Comper WD (1990) ‘Generic’ physical mechanisms of morphogenesis and pattern formation. Development 110:1–18. https://doi.org/10.1242/dev.110.1.1
Newman SA, Forgacs G, Müller GB (2006) Before programs: the physical origination of multicellular forms. Int J Dev Biol 50:289–299. https://doi.org/10.1387/ijdb.052049sn
Nirenberg MW (1963) The genetic code: II. Sci Am 208(3):80–95
Pattee HH (1969) How does a molecule become a message? Dev Biology (Supplement). 3:1–16
Peirce CS ([1867]1868) On a new list of categories. Proc Amer Acad Arts Sci 7: 287–298. https://doi.org/10.2307/20179567
Polanyi M (1967) Life transcending physics and chemistry. Chem Eng News 4535:54–69. https://doi.org/10.1021/cen-v045n035.p054
Polanyi M (1968) Life’s irreducible structure. Sci New Ser 160:1308–1312
Prigogine I, Nicolis G (1967) On symmetry-breaking instabilities in dissipative systems. J Chem Phys 46(9):3542–3550. https://doi.org/10.1063/1.1841255
Prigogine I, Stengers I (1984) Order out of chaos: man’s new dialogue with nature. Bantam Books, Toronto
Purcell EM (1977) Life at low Reynolds number. Am J Phys. 45.3 https://doi.org/10.1119/1.10903
Queiroz J, Emmeche C, Kull K, El-Hani C (2011) The biosemiotics approach in biology: theoretical bases and applied models. In: Terzis G, Arp R (eds) Information and living systems. Philosophical and scientific perspectives. MIT Press, Cambridge, pp 91–129
de Regt HW (2011) Explanation. In: French S, Saatsi J (eds) The continuum companion to the philosophy of science. Continuum Press, London, pp 157–178
Roth S (2014) Kant, Polanyi, and molecular biology. In: Goy I, Watkins E (eds) Kant’s theory of biology. De Gruyter, Berlin, pp 275–291
Salmon WC (1984) Scientific explanation and the causal structure of the world. Princeton University Press, Princeton
Schrödinger E (1992) [1944]. What is life? The physical aspect of the living cell. Cambridge University Press, Cambridge
Sebeok TA (2001) Biosemiotics: its roots, proliferation, and prospects. Semiotica 134 I 4:61–78. https://doi.org/10.1515/semi.2001.014
Shannon CE (1948) A mathematical theory of communication. Bell Syst Tech J 27:379–423
Toffler A (1984) Foreword. Science and change. In: Prigogine I, Stengers I (eds) Order out of chaos: man’s new dialogue with nature. Bantam Books, Toronto, pp xi–xxvi
Trifonov EN (2011) Thirty years of multiple sequence codes. Genomics Proteom Bioinf 9:1–6. https://doi.org/10.1016/S1672-0229(11)60001-6
Walker SI, Davies PCW (2016) The “hard problem” of life. In: Walker SI, Davies PCW, Ellis GFR (eds) From matter to life. Cambridge University Press, Cambridge, pp 19–37. https://arxiv.org/abs/1606.07184v1
Watson JD, Crick FHC (1953) Genetical implications of the structure of deoxyribonucleic acid. Nature 171:964–967
Weaver W ([1949]1964) Recent contributions to the mathematical theory of communication. In: Shannon CE, Weaver W (eds) The mathematical theory of communication. University of Illinois Press, Urbana, pp 1–28
Yockey HP (2005) Information theory, evolution, and the origin of life. Cambridge University Press, Cambridge
Acknowledgments
We thank Dr. Marnix Medema and two anonymous reviewers for their very useful comments on earlier versions of the article.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
Wim Beekman: conceptualization, methodology, investigation, writing original draft.
Henk Jochemsen: conceptualization, supervision, reviewing, and editing.
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no relevant financial or nonfinancial interests to disclose.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Beekman, W., Jochemsen, H. Self-Organization Through Semiosis. Biol Theory 18, 90–100 (2023). https://doi.org/10.1007/s13752-023-00432-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13752-023-00432-6